Abstract
The universal N(6)-threonylcarbamoyladenosine (t6A) modification at position 37 of ANN-decoding tRNAs is central to translational fidelity. In bacteria, t6A biosynthesis is catalyzed by the proteins TsaB, TsaC/TsaC2, TsaD and TsaE. Despite intense research, the molecular mechanisms underlying t6A biosynthesis are poorly understood. Here, we report biochemical and biophysical studies of the t6A biosynthesis system from Thermotoga maritima. Small angle X-ray scattering analysis reveals a symmetric 2:2 stoichiometric complex of TsaB and TsaD (TsaB2D2), as well as 2:2:2 complex (TsaB2D2E2), in which TsaB acts as a dimerization module, similar to the role of Pcc1 in the archaeal system. The TsaB2D2 complex is the minimal platform for the binding of one tRNA molecule, which can then accommodate a single TsaE subunit. Kinetic data demonstrate that TsaB2D2 alone, and a TsaB2D2E1 complex with TsaE mutants deficient in adenosine triphosphatase (ATPase) activity, can catalyze only a single cycle of t6A synthesis, while gel shift experiments provide evidence that the role of TsaE-catalyzed ATP hydrolysis occurs after the release of product tRNA. Based on these results, we propose a model for t6A biosynthesis in bacteria.
INTRODUCTION
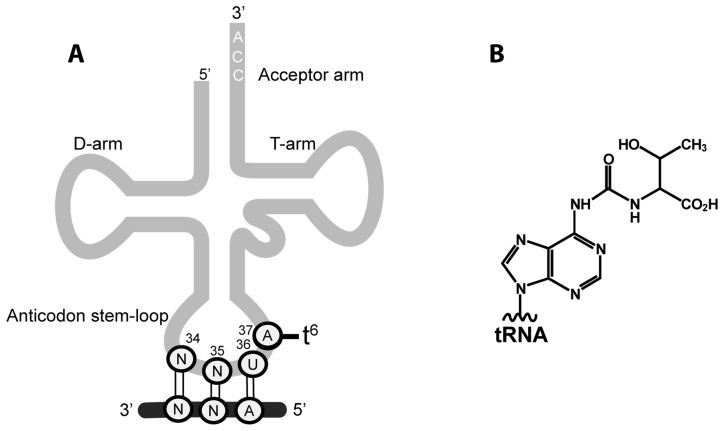
N6-threonylcarbamoyladenosine (t6A) is a universally conserved modified nucleoside found at position 37 in the anticodon stem-loop of many tRNAs decoding ANN codons (N is any nucleotide) in all domains of life (Figure 1). The (t6A37) modification shapes the architecture of the anticodon stem-loop, a requisite step for accurate decoding of mRNA codons (1,2), and is one of the few modifications found in all domains of life (3,4). Genotypes that are defective in t6A37 modification exhibit an increased frequency of frameshift events and inaccurate selection of start codons; these outcomes severely impair normal cellular functions resulting in pleiotropic phenotypes (3,5), neurodegeneration in humans (6) and are implied in the renal-neurological disease Galloway–Mowat syndrome (7). t6A biosynthesis occurs in two steps: members of the TsaC/TsaC2 (formerly named YrdC/Sua5) family, present in all three domains of life (8,9), utilize L-threonine, bicarbonate and adenosine triphosphate (ATP) to synthesize the unstable pathway intermediate threonylcarbamoyl adenylate (TC-AMP) (10). The threonylcarbamoyl (TC) moiety of TC-AMP is then transferred onto A37 of the substrate tRNA by the threonylcarbamoyl transfer (TCT) complex, composed of TsaB, TsaD and TsaE in bacteria (Tsa proteins; e.g. YeaZ, YgjD and YjeE in Escherichia coli (11); and YdiC, YdiE and YdiB in Bacillus subtilis (10)), by the KEOPS complex (Kinase, putative Endopeptidase and Other Proteins of Small size) in archaea (12) and the cytosols of eukaryotes (13,14), and by the TsaD ortholog Qri7 in mitochondria (15). The TsaD/Kae1/Qri7 protein family is responsible for the catalysis of the transfer reaction. The archaeal KEOPS complex is a duplicated linear arrangement of four proteins Cgi121-Bud32-Kae1-Pcc1-Pcc1-Kae1-Bud32-Cgi121 (16) whereas the eukaryotic KEOPS complex (studied so far only in yeast and human) is a linear complex of Gon7-Pcc1-Kae1-Bud32-Cgi121 (17,18), and Qri7 is a quarantined dimer (15).
Figure 1.
Location and structure of t6A37 in tRNA. (A) Schematic of the anticodon stem-loop of tRNANNU and the mRNA ANN codon it decodes. (B) The chemical structure of t6A.
The t6A biosynthesis pathway is essential in bacteria (an exception is Streptococcus mutans (19)) and TsaE and TsaB are bacterial specific, establishing these proteins as compelling new targets for antimicrobial development. Further, an essential interaction network between TsaB, TsaD and TsaE in vivo has been well documented (20–22). In vitro pull-down assays on the purified E. coli proteins showed that TsaC forms complexes with either TsaD or TsaB, while neither protein alone bound to TsaE (11). Later, the crystal structures of the heterodimer of TsaD and TsaB from E. coli (23) and Salmonella typhimurium (24) were determined, and a 1:1:1 stoichiometric complex of E. coli TsaB, TsaD and TsaE was isolated and its solution structure determined by small angle X-ray scattering (SAXS) (23). ATP-mediated formation of this complex is reportedly required for t6A biosynthesis on tRNA (23). In S. typhimurium, isothermal titration calorimetry (ITC) experiments revealed ATP- and adenosine diphosphate (ADP)-dependent binding of TsaE to the TsaB/TsaD 1:1 complex, and an ATP dependent binding of TsaE to TsaD (24). TsaE is an atypical ATPase that hydrolyzes ATP to ADP only in the presence of TsaB and TsaD (11), and this ATPase activity is necessary for cell growth (25).
Although the subunit interactions in several bacterial t6A biosynthesis systems have been elucidated, their exact roles in the catalytic cycle and in recognition of the tRNA substrate remain unknown. Furthermore, the contribution of the hydrolysis of ATP by TsaE to the biosynthesis of t6A in bacteria remains unclear. Here we present biophysical, biochemical and kinetic studies of the structure and mechanism of the Thermotoga maritima t6A biosynthesis system. The results show that a larger subcomplex, formed by the subunits TsaB and TsaD and possessing a dimeric structure, is the tRNA binding scaffold and reveal the role of TsaE and its ATPase activity to be needed after the turnover and dissociation of the modified tRNA product.
MATERIALS AND METHODS
Cloning procedures
The T. maritima tsaD gene (NCBI accession number WP_004082750) was synthesized by Genescript (www.genescript.com) and cloned into the BamHI (5′-end) and Xho1 (3′-end) sites of pET-45b (Novagen, San Diego, CA) in-frame with a non-cleavable N-terminal His6 tag. Clones of T. maritima tsaB (NCBI accession number WP_004080725), tsaC2 (NCBI accession number AGL49779), and tsaE (NCBI accession number AGL50565) in a pBAD vector were kindly provided by the Joint Center for Structural Genomics (La Jolla, CA, (26)) and were subcloned into the NheI (5′-end) and HindIII (3′-end) restriction sites of a pET-28a expression vector (Novagen, San Diego, CA) in-frame with the N-terminal His6 tag and a thrombin cleavage site. The nucleotide sequences of the resulting constructs, pET28a-tsaB, pET28a-tsaC2, pET28a-tsaE and pET45B-tsaD, were confirmed by sequencing (Genewiz, Inc., South Plainfield, NJ, USA) and used for expression and purification. All primers used for cloning are listed in Supplementary Table S1.
Single-site mutagenesis of T. maritima TsaE
Point mutagenesis was carried out on TsaE (cloned in pET-28a) using the Q5® Site-Directed Mutagenesis Kit (NEW ENGLAND Biolabs Inc.). The Glu108 residue was mutated to Ala (GAA→GCA). Thr43 was mutated to Ala (ACG→GCA). Primers used for the mutations are listed in Supplementary Table S1. Resulting DNA constructs were confirmed to be correct and in the frame by sequencing (at Genewiz, Inc, South Plainfield, NJ, USA).
Protein expression and purification
All constructs of the T. maritima wild-type and mutant proteins used in this study were expressed in E. coli Overexpress™ C41 (DE3) cells (Lucigen Corporation, Middleton, WI, USA). Cultures were grown in LB-Miller broth containing kanamycin (50 μg/ml) at 37°C with shaking. Expression was induced at A600 = ∼0.6 by addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM, followed by 3–4 h of growth. Cells were harvested by centrifugation at 6000 × g for 15 min at 4°C, and the resulting cell pellets were suspended and lysed in 40 ml of lysis buffer containing 50 mM Tris (pH 7.5), 100 mM NaCl, 10% glycerol and one cOmplete™ ULTRA tablet of protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IL, USA). The cell lysates were centrifuged at 20 000 × g for 30 min at 4°C, and filtered using a 0.22 μm filter before being loaded onto a 5 ml Ni–NTA (Qiagen, Valencia, CA, USA) column pre-equilibrated with binding buffer containing 50 mM Tris (pH 7.5), 100 mM NaCl, 10 mM imidazole and 1 mM β-mercaptoethanol (BME) (Buffer A). For purification of TsaC2 and TsaE, the column was washed with 25 ml of buffer A, followed by a wash with 100 ml of wash buffer containing 50 mM Tris, 500 mM NaCl, 20 mM imidazole (pH 7.5) and 1 mM BME (buffer B). The His6 tag was removed by thrombin digestion (100 units) of the Ni-NTA-bound proteins for 10–16 h at 22°C. For purification of TsaB and TsaD, the proteins were eluted from the Ni-NTA resin with buffer A supplemented with 300 mM imidazole after thorough washing with buffer B. For all proteins, elution fractions were pooled and subjected to size-exclusion chromatography using a Sephacryl S-200 HR 16/60 column (GE Healthcare Life Sciences, Pittsburgh, PA, USA) pre-equilibrated with 50 mM Tris (pH 7.5), 100 mM NaCl and 1 mM dithiothreitol (DTT). For all proteins, purity of >95% was verified by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). Escherichia coli TsaB, TsaD and TsaE were overexpressed and purified as described previously (11).
Native gel migration assay
For native gel analysis of the interactions of T. maritima Tsa proteins, protein combinations were mixed in a final volume of 20 μl, with each protein component set at a final concentration of 10 μM, in buffer containing 25 mM Tris–HCl (pH 7.5), 50 mM NaCl, 1 mM DTT and 5% glycerol. A total of 20 μl were loaded onto a 1.2% agarose gel, and electrophoresed for 50 min at 135 mV on ice in a running buffer composed of 30 mM Tris–HCl (pH 7.5) and 100 mM glycine. Proteins were visualized on the gel by Coomassie Blue staining.
Size exclusion chromatography
Gel filtration experiments were carried out using an Enrich SEC 650 10 × 300 mm column (BIO-RAD, Hercules, CA, USA) and an NGC chromatography system (Bio-Rad). The column was calibrated as described in Supplementary Methods. A total of 500 μl of sample were loaded on the column pre-equilibrated in buffer containing 50 mM Tris–HCl (pH 7.5), 50 mM NaCl and 1 mm DTT and eluted using the same buffer at a flow rate of 0.25 ml/min, with detection at 280 nm. A total of 1 mM ATP was included in the buffer used for isolation of the protein heterocomplexes.
Small angle X-ray scattering (SAXS) data acquisition and analysis
SAXS data were collected on the Bio-SAXS beamline BL4–2 at the Stanford Synchrotron Research Laboratory (27) using a Rayonix MX225-HE CCD detector (Rayonix, Evanston, IL, USA) with a sample-to-detector distance of 1.7 m and a beam energy of 11 keV (λ = 1.127 Å). The momentum transfer (scattering vector) q was defined as q = 4π sin(θ)/λ, where 2θ is the scattering angle. The q scale was calibrated by silver behenate powder diffraction (28). All data were collected to a maximum q of 0.53 Å−1. For the TsaB2D2 complex, scattering data were collected at a wavelength of 1.3 Å for 10 consecutive 2-s exposures from a sample containing 1.1 mg/ml protein, 50 mM Tris (pH 7.5), 100 mM NaCl and 1 mM DTT. Scattering data were collected from the buffer alone and subtracted from the total protein solution scattering. For the TsaB2D2E2 complex, a 100 μl sample containing 20 μM of each of TsaB and TsaD and 80 μM TsaE was prepared in buffer containing 50 mM Tris–HCl (pH 7.5), 100 mM NaCl, 1 mM DTT and 1 mM ATP; and incubated for 30 min. The sample was loaded onto a Superdex 200 PC 3.2/30 column (GE Healthcare Life Sciences) pre-equilibrated in the same buffer. The protein was eluted from the column at a flow rate of 0.05 ml/min and passed through a 1.5-mm quartz capillary cell (Hampton Research, Aliso Viejo, CA, USA) at 20°C in line with the X-ray beam. Scattering images were obtained with 1-s exposures every 5 s using the data acquisition program Blu-ICE (29,30). The data processing program SasTool (27) was used for scaling, azimuthal integration and averaging of individual scattering images after inspection for any variations potentially caused by radiation damage. The first 100 frames were scaled and averaged to create a buffer-scattering profile, which was then subtracted from each of the subsequent images to produce the final scattering curve for each frame. Data analysis was performed using tools in the ATSAS package (31). Data for every five frames along the course of the elution profile and free of signs of sample aggregation were averaged using PRIMUS (32). Radii of gyration (Rg), extrapolated from the Guinier region of the Guinier plot, were computed using PRIMUS (32). P(r) functions were calculated using the program GNOM (33). Theoretical scattering curves were computed from different structural models and compared to experimental scattering curves using the program FoXS (34). The relative Porod volume was determined from the SAXS data using DATPOROD, and the molecular mass was determined using DATMOW (31) and SAXS-MoW (HTTP://www.if.sc.usp.br/∼saxs/). Normal mode analysis (NMA) was performed using the program Sreflex (35) and conformational sampling and multistate modeling were executed in MultiFoXS (36). Ab initio envelop reconstruction was performed using the programs DAMMIN (37) and DAMAVER (38) in the ATSAS software package (31). The structural parameters derived from the SAXS data are summarized in Table 1.
Table 1. Parameters calculated from SAXS data.
| TsaB2D2 | TsaB2D2E2 | |
|---|---|---|
| Structural parameters | ||
| Rg (Å) from Guinier plot | 44.20 ± 0.06 | 48.22 ± 0.07 |
| Rg (Å) from P(r) | 44.86 ± 0.08 | 47.84 ± 0.1 |
| Dmax (Å) | 148.9 | 150 |
| Porod volume estimate, Vp (Å3) | 250,783 | 289,003 |
| Molecular mass (kDa) | ||
| From DATMOW | 129.22 | 158.85 |
| From DAMMIN models | 121.35 | 141.35 |
| From theoretical protein sequence | 128 | 166 |
| Ab initio modeling parameters | ||
| DAMAVER NSD (averaged over 10 models)a | 0.532 ± 0.142 | 0.69 ± 0.04 |
aNSD: normalized spatial discrepancy.
3D homology modeling
Homology models of T. maritima TsaD and TsaE were generated using the ModWeb server (https://modbase.compbio.ucsf.edu/modweb/) (39), and any missing residues were built using Modeller (40). To build a homology model of the T. maritima TsaB2D2 stoichiometric complex, the crystal structure of the T. maritima TsaB homodimer (form-II) (41) was used as the core structure (after building any missing residues in Modeller), and the positions of the two TsaD subunits were determined based on the position of the TsaD subunit in the crystal structure of the heterodimeric complex of E. coli TsaD and TsaB (YgjD and YeaZ) (PDB ID: 4YDU (23)) by superposing the TsaB subunit of the E. coli complex onto each protomer of the crystal structure of the T. maritima TsaB homodimer. Subsequently, two copies of the monomeric homology model of T. maritima TsaD were put in place of the E. coli TsaD subunits by superpositioning to yield the final TsaB2D2 model. To generate a structural model of the TsaB2D2E2 stoichiometric complex, two copies of the homology model of T. maritima TsaE were docked onto the model of TsaB2D2 using ZDOCK (42). Docking solutions were ranked based on chi values against experimental SAXS data and electrostatic and surface complementarity between each TsaE subunit and a concave surface on the TsaB2D2 complex. However, during the course of this study, we obtained crystals of the TsaB2D2E2 complex and collected low resolution data (5 Å) and the preliminary crystallographic electron density maps revealed a more accurate orientation of the TsaE subunits in the complex (Supplementary Figure S1). This information was used to improve our homology model of TsaB2D2E2 which was then used for fitting to the SAXS data. All structural alignments were performed in Coot (43) and structure figures were prepared in PyMOL (Version 1.8 Schrödinger, LLC).
tRNA transcription and purification
Escherichia coli tRNAThrCGU and E. coli tRNALysUUU, used for tRNA binding studies and radiochemical incorporation assays, respectively, were produced by in vitro transcription as detailed in Supplementary Methods.
Electrophoretic mobility shift assay for tRNA binding studies
The binding reactions were carried out in 10-μl solutions containing 17 nM (0.07 pmol) 32P-labeled E. coli tRNAThr, 0.25–10 μM purified recombinant proteins or protein complexes, 50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 5% glycerol, 1 mM DTT, 0.1–0.5 mM ATP and 1 unit of RNasin®. For the tRNA and TsaE competition assay, the binding reactions contained 5 μM T. maritima TsaB2D2 complex pre-incubated with 100 nM 32P-labeled E. coli tRNAThr for 15 min, added 1–15 μM T. maritima TsaE and the same buffer conditions as above. For determination of the tRNA binding stoichiometry, the reactions contained 25 μM unlabeled E. coli tRNAThr, 100 nM 32P-labeled E. coli tRNAThr and 6–75 μM pre-isolated T. maritima TsaB2D2 complex in the same buffer conditions as above. Following incubation for 15 min at 4°C, the reactions were loaded onto a 10% non-denaturating polyacrylamide gel and electrophoresis was performed in 1× Tris-Glycine-EDTA (TGE) buffer at 4°C for 3 h at 7.5 V/cm. The gels were autoradiographed to visualize the tRNA and—when applicable—stained with Coomassie Blue to visualize the proteins. Band densities were measured using Image Studio™ Lite 5.x (LI-COR Biotechnology, Lincoln, NE, USA), and the density data were fit using the nonlinear least squares method (44) in the program GraphPad Prism version 7.00 (GraphPad Software, La Jolla California USA, www.graphpad.com).
Enzymatic activity assays for formation of t6A-modified tRNA (radiochemical incorporation assay)
Initial enzyme assays to test the ability of various T. maritima Tsa proteins to synthesize t6A-modified tRNA were performed in a volume of 50 μl containing 100 mM Tris (pH 8.0), 300 mM KCl, 5.0 mM DTT, 2 mM ATP, 20 mM MgCl2, 20 μM tRNALysUUU, 50 mM NaHCO3, 50 μM L-[U-14C]-threonine (100 000 DPM/assay) and 10 μM TsaB, TsaC2, TsaD and TsaE. Assays were incubated at 60°C for 1 h, and the RNA was precipitated by the addition of 500 μl 10% TCA and cooling on ice for at least 10 min. RNA was then collected by filtration through Whatman GF/C filters, and the filters were subsequently washed with 100% cold ethanol. The dried filters were combined with Econo-Safe scintillation cocktail (RPI™) and counted in a Hidex 300-SL liquid scintillation counter.
Enzyme time-course assays measuring the incorporation of L-[14C]-threonine into RNA transcripts over time were performed in a total volume of 225 μl containing 100 mM Tris (pH 7.5), 300 mM KCl, 5.0 mM DTT, 2 mM ATP, 20 mM MgCl2, 20 μM tRNALysUUU, 50 mM NaHCO3, 250 μM L-[14C]-threonine (4 500 000 DPM/assay), 4 μM each TsaB, TsaC2, and TsaD, and 2 μM TsaE, TsaET42A, or TsaEE108A (where appropriate). Assays were incubated at 37°C for 2 h and 50 μl aliquots were taken at 10, 30, 60 and 120 min. For assays with variable [TsaE], the TsaB, TsaC2, TsaD proteins were present at 2 μM each, and TsaE was varied from 0.4 to 10 μM. Assays were incubated at 37°C for 1 h and 50 μl aliquots were taken at 10, 30 and 60 min. RNA was precipitated and counted as described above.
Isothermal titration calorimetric (ITC) assay for protein-protein and protein-nucleotide binding
ITC measurements of the thermodynamic parameters of protein–protein and protein–ATP binding equilibria were performed with a MicroCalITC200 instrument (GE Healthcare). For each measurement, a total of 20 injections of 2 μl of the injectant protein were performed at intervals of 120 s under continuous stirring. Normalized area data in kcal/mol of integrated peaks were plotted versus the molar ratio of injectant to sample titrated, using the Origin 7.0 software (OriginLab Corporation, Northampton, MA, USA).
ATP binding measurements by MANT fluorescence
For qualitative assessment of the ability of wild-type T. maritima TsaE and mutants TsaEE108A and TsaET42A to bind ATP upon association with TsaB2D2, the fluorescently labeled β,γ non-hydrolyzable ATP analog 2′/3′-O-(N-Methyl-anthraniloyl)-adenosine-5′-[(β,γ)-imido] triphosphate (MANT-AMPPNP) (Jena Bioscience, Farmingdale, NY, USA) was employed. MANT-AMPPNP was added to 200-μl samples containing 1 μM pre-isolated TsaB2D2 and 1 μM wild-type or mutant TsaE proteins (thus forming the respective TsaB2D2E1 complexes) in buffer containing 25 mM Tris (pH 7.6), 50 mM KCl and 2 mM MgCl2. Additionally, samples contained 1 μM TsaB2D2 and 2 μM TsaE or mutant proteins (thus forming the respective TsaB2D2E2 complexes). The final nucleotide concentration was 0.1 μM. Nucleotide binding was detected by monitoring the increase in MANT fluorescence (λex = 355 nm, λem = 448 nm), relative to its fluorescence in the absence of proteins, using a Fluoromax-3 spectrofluorimeter (Horiba Scientific, Edison, NJ, USA). As controls, pre-isolated TsaB2D2 alone and individual proteins were similarly tested in this assay.
TsaE ATPase activity assay
The ATP hydrolysis was carried out at 37°C in 20-μl reactions containing 5 μM wild-type or mutant TsaE with or without an equimolar concentration of TsaB2D2, 49 μM ATP, 1 μM [γ-32P] ATP (PerkinElmer), 25 mM Tris (pH 7.6), 50 mM KCl and 2 mM MgCl2. The reactions were stopped by the addition of 0.5 μl of 10% SDS. The 2 μl of each reaction were spotted on a PEI–cellulose TLC plate (Sigma-Aldrich) and radioactive nucleotides were separated using 0.5 M KH2PO4 (pH 3.5) as solvent. Plates were dried, and radioactivity was revealed by phosphorimaging.
Native PAGE analysis of t6A enzymatic assays
For native gel analysis of the interaction of product tRNA with components of the T. maritima TCT complex, reactions were performed in a volume of 50 μl and contained 100 mM Tris (pH 7.5), 300 mM KCl, 5 mM DTT, 50 mM sodium bicarbonate, 20 mM MgCl2, 50 μM [U-14C]threonine (1 000 000 dpm), 2 mM ATP, 5% Glycerol, 8 μM each TsaC2, TsaB, TsaD and 4 μM either TsaE or TsaEE108A where indicated. The tRNA concentration was set at 40 μM except in a single reaction with TsaB and TsaD, in which case the tRNA concentration was at 3.5 μM. Reactions were incubated at 37°C for 1 h.
To obtain an independent sample of [14C]-labeled t6A-modified tRNA, a 100 μl reaction containing the components above was carried out at 37°C for 1 h. The assay mixture was diluted to 400 μl with water, and 40 μl of 7.5 M ammonium acetate was added. A mixture of phenol:chloroform:isoamyl alcohol (25:24:1; 440 μl) was added and, after vortexing, the aqueous layer was transferred to 900 μl of cold 100% ethanol. The mixture was stored at −20°C for 30 min, subsequently centrifuged at 16 300 g for 30 min, the supernatant removed and the tRNA pellet re-suspended in 50 μl Tris–EDTA buffer (pH 7.5).
A 20 μl aliquot of each reaction above, and a 5 μl aliquot of [14C]-labeled tRNA, were loaded onto a 10% non-denaturing polyacrylamide gel and electrophoresis was performed in 1× TGE buffer at 4°C for 2 h at 10 mA. The gel was fixed (40% methanol, 10% acetic acid) for 1 h and exposed to a phosphorimager screen overnight. The screen was scanned using a GE Healthcare Typhoon Trio Variable Mode Imager Scanner, and the scan visualized using ImageQuant TL.
RESULTS
Cloning of tsaB/C2/D/E genes from T. maritima and over-production and purification of the recombinant proteins
The T. maritima tsa genes were obtained from the Joint Center for Structural Genomics clone collection (26) and sub-cloned into pET-28a or pET-45b vectors, with either a cleavable (in the case of TsaB/C2/E) or non-cleavable (in the case of TsaD) N-terminal His6 tag. Individual proteins were overexpressed in E. coli and purified to homogeneity by Ni-NTA and gel filtration chromatographies. The His6 tag was successfully cleaved from all proteins except TsaD. Attempts to produce TsaD with a cleavable His6 tag failed due to solubility problems (data not shown). Purity was verified by SDS-PAGE (Supplementary Figure S2A).
In vitro reconstitution of t6A37 forming activity of T. maritima Tsa proteins
The in vitro formation of t6A37 by T. maritima TsaB, TsaC2, TsaD and TsaE was probed using radiochemical-based assays in which the incorporation of [14C]threonine into the RNA substrate was measured. As previously observed with the E. coli t6A37 system (11), robust incorporation of radioactivity into the tRNA transcript was observed only in assays containing all four proteins (Supplementary Figure S2B).
Interactions of T. maritima Tsa proteins and isolation of stable TsaB2D2E1 and TsaB2D2E2 complexes
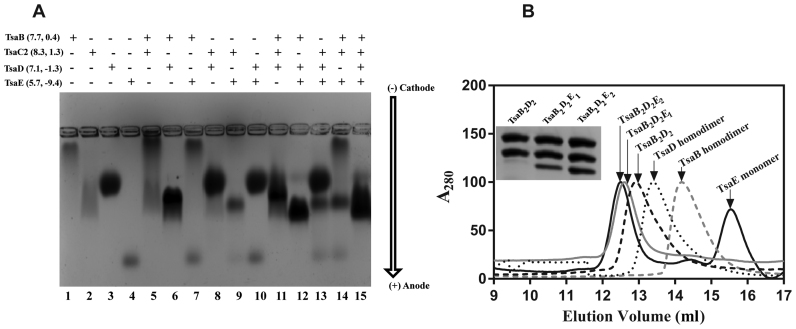
To investigate the interactions between the four Tsa proteins of T. maritima, we analyzed their equimolar binary, ternary and quaternary mixtures by native agarose gel electrophoresis (Figure 2A). The results show formation of a TsaB/TsaD complex (Figure 2A, lane 6) and a TsaB/TsaD/TsaE complex (Figure 2A, lane 12), as previously observed for the E. coli (23) and S. typhimurium (24) orthologs. TsaE does not interact with individual TsaB or TsaD proteins (Figure 2A, lanes 7 and 10). However, TsaE forms a complex with TsaC2 as evidenced by almost complete disappearance of the TsaE band, accompanied by narrowing of the TsaC2 band on the gel (Figure 2A, lane 9). Further, the TsaE/TsaC2 heterocomplex is formed even in the presence of TsaD (Figure 2A, lane 13) or TsaB (Figure 2A, lane 14). Mixing of all four proteins gives rise to a broad band with mobility spanning that of the TsaB/TsaD, TsaB/TsaD/TsaE and TsaC2/TsaE complexes (Figure 2A, lane 15), suggesting the presence of all three complexes in the ternary mixture.
Figure 2.
Identification and isolation of three stable complexes of Thermotoga maritima Tsa proteins. (A) Native agarose gel electrophoresis analysis of individual T. maritima Tsa proteins (lanes 1–4) and various equimolar mixtures (lanes 5–15). (B) Size-exclusion chromatographic (SEC) profiles of individual TsaB (gray dashed line) and TsaD (dotted line) proteins, of the TsaB2D2 complex (black dashed line) formed from an equimolar mixture of TsaB and TsaD, of the TsaB2D2E1 complex (gray solid line) formed from an equimolar mixture of TsaB, TsaD and TsaE, and of the TsaB2D2E2 complex (black solid line) formed by adding excess TsaE to an equimolar mixture of TsaB and TsaD. All SEC profiles were collected by monitoring absorbance at 280 nm. Inset: SDS-PAGE analysis of SEC fractions corresponding to the major SEC peaks for the protein mixtures.
We then used size-exclusion chromatography to isolate the different complexes observed by the gel shift assay. For isolation of homocomplexes of individual proteins, no ATP was included in the sample or chromatography buffer. For isolation of heterocomplexes of mixed proteins, 1 mM ATP was added to sample and buffer. Chromatography data confirm that TsaB and TsaD each elute as a homodimeric complex with a homodimer molecular mass of 48 and 78 kDa, respectively (Figure 2B), consistent with the known homodimerization of the orthologs from E. coli (23), S. typhimurium (45), Vibrio parahaemolyticus (46), Pseudomonas aeruginosa (47) and with the crystallographically observed homodimer of T. maritima TsaB itself (41). Upon equimolar mixing of TsaB and TsaD, they form a complex with an apparent mass of 120 kDa (Figure 2B), consistent with a 2:2 stoichiometric complex (TsaB2D2). Chromatographic analysis of an equimolar mixture of TsaB, TsaD and TsaE in the presence of ATP yields a single peak corresponding to an apparent molecular mass of 141 kDa, consistent with a TsaB/TsaD/TsaE complex with a subunit stoichiometry of 2:2:1 (TsaB2D2E1). However, when 1–3 molar excess of TsaE is included in the mixture, a larger, stable TsaB/TsaD/TsaE complex can be isolated with an apparent mass of 158 kDa, consistent with a subunit stoichiometry of 2:2:2 (TsaB2D2E2). SDS-PAGE analysis of the peak fractions confirms the subunit contents and is consistent with the deduced stoichiometries of the three complexes (Figure 2B and Supplementary Figure S3). Further, as previously shown in the E. coli system (23), size-exclusion chromatography demonstrates that TsaE does not bind to either TsaB or TsaD homodimer (Supplementary Figure S4), indicating that both TsaB and TsaD are necessary to create the TsaE binding site (23).
To investigate whether the association of TsaE with TsaB2D2 is ATP-mediated, as previously reported with the E. coli (23) and S. typhimurium (24) systems, we measured the affinity of TsaE to TsaB2D2 in the absence and presence of nucleotides and MgCl2 by ITC. Titration of TsaE into TsaB2D2, first in the absence of nucleotides then in the presence of ADP, resulted in binding with Kd = 1.3 ± 0.001 μM and 1.1 ± 0.1 μM, respectively (Table 2 and Supplementary Figure S5), whereas titration in the presence of 1 mM non-hydrolyzable ATP analog AMPPCP and MgCl2 yielded a Kd of 0.34 ± 0.07 μM. These results show that, unlike the E. coli and S. typhimurium systems, T. maritima TsaE is able to bind to TsaB2D2 in the absence of nucleotides, although affinity is enhanced approximately 4-fold in the presence of ATP but not in the presence of ADP.
Table 2. The thermodynamic parameters measured by ITC for the association of TsaE and mutants to TsaB2D2.
| Titrant (μM) | Injectant (μM) | Stoichiometry (n) | K d (μM) | ΔH (kcal/mol) | ΔS (kcal/mol/deg) |
|---|---|---|---|---|---|
| TsaB2D2 (20)a | TsaE (400) | 2.72 | 0.34 ± 0.07 | 7.44 ± 0.13 | 54.7 |
| TsaB2D2 (20)b | TsaE (400) | 2.16 | 1.1 ± 0.1 | 1.54 ± 0.054 | 32.5 |
| TsaB2D2 (20) | TsaE (400) | 2.11 | 1.3 ± 0.001 | 11.9 ± 0.072 | 67.2 |
| TsaB2D2 (20)a | TsaET42A (400) | 2.0 | 1.1 ± 0.01 | 10.34 ± 0.14 | 62.2 |
| TsaB2D2 (20) | TsaET42A (400) | 2.8 | 1.4 ± 0.001 | 8.0 ± 0.62 | 53.9 |
| TsaB2D2 (20)a | TsaEE108A (400) | 1.91 | 2.4 ± 0.02 | 11.90 ± 0.17 | 65.9 |
| TsaB2D2 (20) | TsaEE108A (400) | 2.9 | 1.1 ± 0.001 | 9.3 ± 0.38 | 59.1 |
ain the presence of 1 mM AMPPCP and MgCl2.
bin the presence of 1 mM ADP and MgCl2.
SAXS structure of TsaB2D2 reveals a symmetric dimer of heterodimers with flexible inter-subunit interfaces
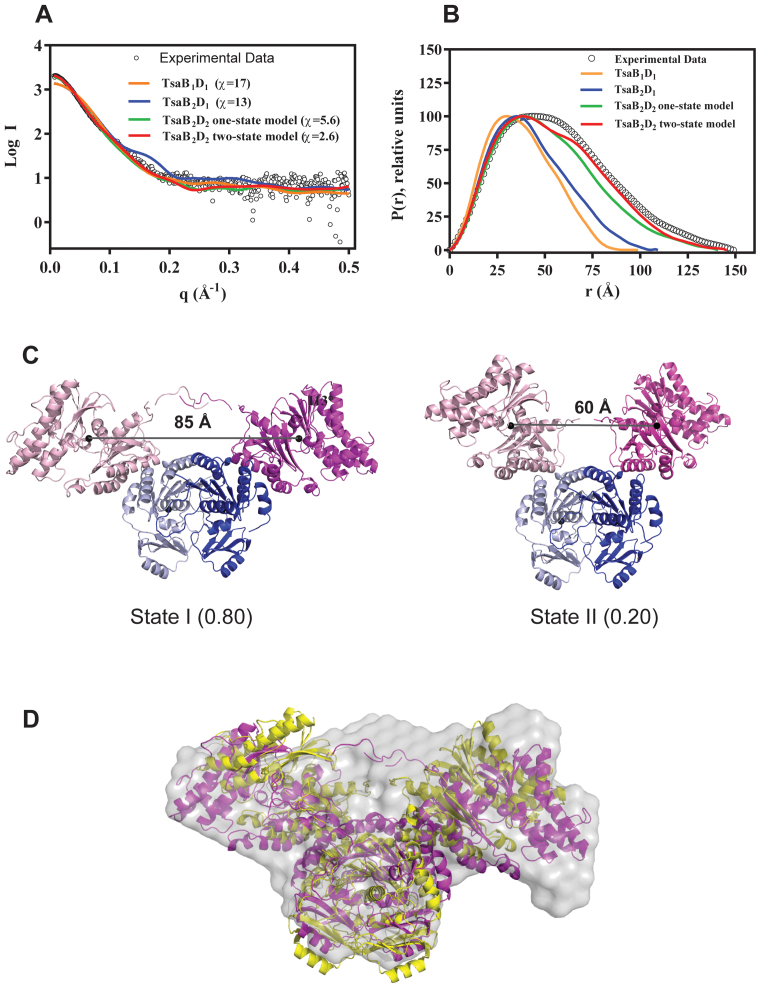
To confirm the stoichiometries and obtain structural data of the complexes in solution, we performed SAXS experiments on the TsaB2D2 and TsaB2D2E2 complexes purified by gel filtration. Structural parameters derived from the SAXS data are listed in Table 1. For the TsaB2D2 complex, theoretical scattering curves and intra-particle distance distribution functions (P(r)) were calculated from 3D homology models representing hypothetical 1:1, 2:1 and 2:2 stoichiometric complexes of TsaB and TsaD (generated as described in ‘Materials and Methods’ section), and compared to the experimental scattering curve and P(r) function. Although the TsaB2D2 homology model provides the best fit to the experimental scattering data (χ = 5.6) (Figure 3A and B), a loss of plateau in the Porod–Debye plot (Supplementary Figure S6) and the discrepancy between the theoretical Rg (40.44 Å) and the experimental Rg (44.2 Å) suggest structural plasticity in the TsaB2D2 complex in solution. We therefore tested if a weighted mixture of multiple models better represents the solution scattering. NMA, followed by conformational sampling and multistate modeling resulted in a two-state model constituted of two conformations with a distribution of 80%:20%. This model provides a significantly improved fit to the experimental scattering data (χ = 2.6) and P(r) distribution (Figure 3A and B), as well as a calculated model Rg of 43 Å, in agreement with the experimental value. In this model, the TsaD subunits are mobile and their interfaces with the TsaB subunits are loose, as indicated by a marked difference between the two observed conformations. The major conformer is an open form of TsaB2D2, exhibiting a distance between the centers of mass of the two TsaD subunits of 85 Å (Figure 3C). In the minor conformer, which represents a closed form, this distance is 60 Å. Ab initio shape reconstruction from the SAXS data, using 10 independent models and without enforcing 2-fold symmetry, yielded an average model with a reasonable normalized spatial discrepancy (NSD) of 0.532. The NMA-optimized two-state model shows an excellent fit to the ab initio molecular envelope (Figure 3D).
Figure 3.
The SAXS solution structure of the TsaB2D2 complex. (A) Comparison of the solution scattering curve (black circles) and the theoretical scattering curves calculated from various attempted models (colored lines), and associated chi values. (B) Comparison of the P(r) functions calculated from the experimental data (black circles) and the theoretical scattering curves (colored lines), showing best fit to the two-state model. (C) Ribbon diagrams of the two states of the TsaB2D2 complex in the two-state model. The TsaB and TsaD subunits are colored in two shades of blue and magenta, respectively. The numbers in parentheses represent the relative abundances of the states in solution. The distanced between the centers of mass of the TsaD subunits are shown. (D) Ab initio reconstruction of the molecular envelope of the TsaB2D2 complex (gray surface) calculated from the SAXS data and overlaid on states I and II in magenta and yellow, respectively.
SAXS structure of TsaB2D2E2 complex and stabilizing effect of the TsaE subunits
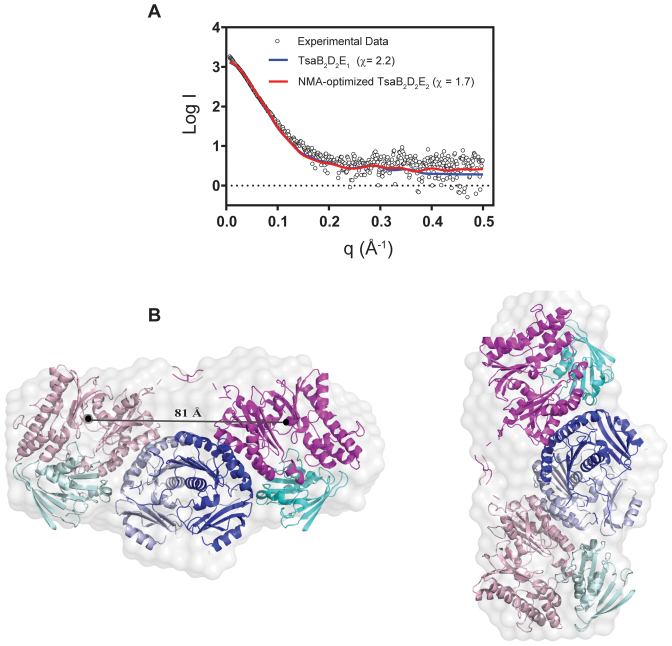
To study the structure of the TsaB2D2E2 complex in solution, SAXS data were collected by employing an FPLC gel filtration step coupled to the SAXS measurements. The Porod volume and molecular mass deduced from the SAXS data (Table 1) are consistent with a monodisperse sample. Further, the experimental Dmax values for TsaB2D2E2 and TsaB2D2 are approximately the same (∼150 Å), indicating that the TsaB2D2E2 structure is not more extended than that of TsaB2D2, consistent with TsaE binding at the interface of TsaB and TsaD. NMA fitting results in a single structural model representing a single conformation of the TsaB2D2E2 complex. The theoretical scattering curve from this model is in good agreement with the experimental scattering curve (χ = 1.7, Figure 4A). At last, ab initio shape reconstruction from the SAXS data, using 10 independent models without enforcing the 2-fold symmetry of the complex, resulted in an average model with NSD value of 0.69 ± 0.04). The NMA-optimized model shows an excellent fit to the ab initio molecular envelope (Figure 4B).
Figure 4.
The SAXS solution structure of the TsaB2D2E2 complex. (A) Comparison of the solution scattering data (black circles) to the theoretical scattering curves calculated from various attempted structural models (colored lines). The chi values for the various models are listed. (B) The averaged ab initio reconstruction of the TsaB2D2E2 molecular envelope calculated from the SAXS data and overlaid on the ribbon diagram of the NMA-optimized model. The TsaB, TsaD and TsaE subunits are colored in two shades of blue, magenta and cyan, respectively. The distance between the centers of mass of the two TsaD subunits is shown.
The TsaB2D2 complex constitutes the minimal platform for tRNA binding
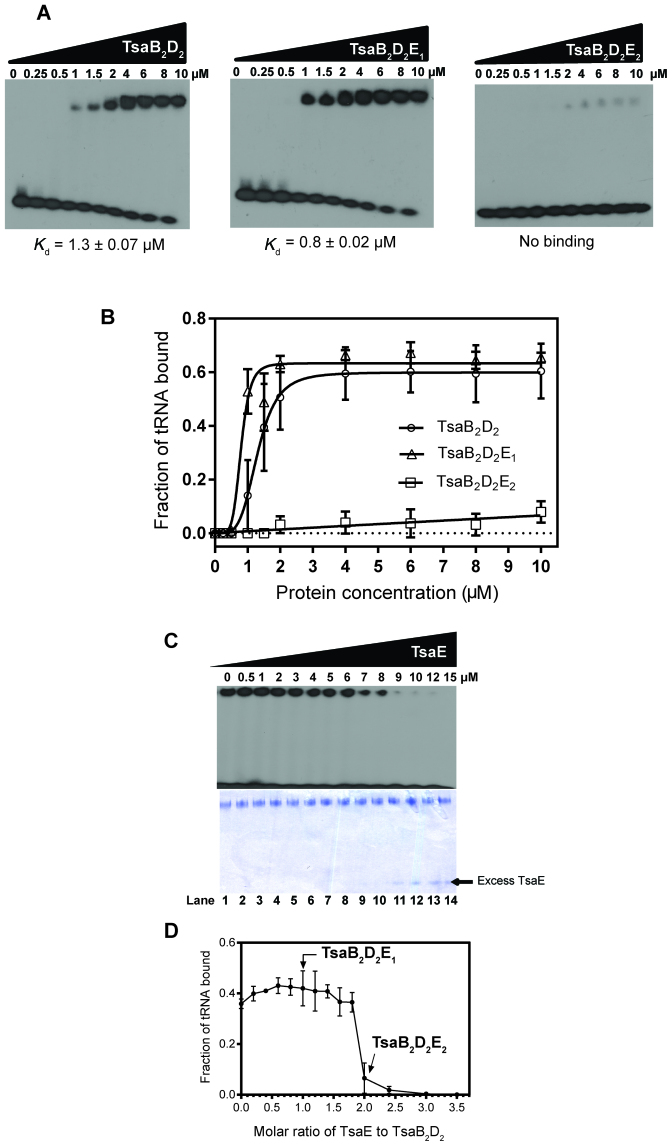
We conducted electrophoretic mobility shift experiments to investigate the binding of individual TsaB, TsaC2, TsaD and TsaE proteins and their complexes (pre-isolated by gel filtration) to 32P-labeled E. coli tRNAThr (we have shown above that E. coli tRNA is a good substrate for the T. maritima enzyme system). Using individual proteins, no nucleoprotein complexes could be detected on the gel, even at high protein concentrations, but weak binding to TsaC2 was observed (Supplementary Figure S7). Unlike the individual proteins, the TsaB2D2 and TsaB2D2E1 complexes bound tRNA with an apparent Kd of 1.3 ± 0.07 and 0.8 ± 0.02 μM, respectively (Figure 5A and B). In contrast, the TsaB2D2E2 complex did not bind tRNA. These data demonstrate that TsaB2D2 is the minimal tRNA binding platform, and indicate that binding of one TsaE subunit to the TsaB2D2 platform maintains, if not slightly increases, its tRNA binding affinity whereas binding of the second TsaE subunit abolishes tRNA binding.
Figure 5.
Binding of tRNAThr by the TsaB homodimer, TsaD homodimer, TsaE and pre-isolated complexes. Proteins were incubated with 100 nM of 32P-labeled tRNAThr at room temperature and the mixtures separated by native PAGE (see ‘Materials and Methods’ section). (A) Autoradiograms showing unbound tRNAThr (at the bottom of each gel) and the nucleoprotein complexes (at the top of each gel). (B) Plot of fraction of tRNA bound quantified by densitometry as the ratio of upper-band intensity to total intensity in each lane. (C) TsaE-dependent release of tRNA from the TsaB2D2 complex. TsaB2D2 complex (5 μM) pre-incubated with 32P-labeled tRNAThr (100 nM) at room temperature was titrated with increasing concentrations of TsaE, and analyzed by native PAGE. Top panel: autoradiogram of the native gel showing free and bound tRNA at the bottom and top of the gel, respectively. Bottom panel: coomassie staining of the same gel. TsaE concentrations are indicated at the top, and lane numbers are indicated at the bottom. (D) Densitometric quantification of the radioactive bands in panel A, n = 2.
To investigate this phenomenon more quantitatively, we performed a competition electrophoretic mobility shift assay (EMSA) experiment in which we probed the ability of TsaE to displace bound tRNA from the TsaB2D2 complex, by adding increasing concentrations of TsaE to TsaB2D2 that is pre-bound to 32P-labeled tRNA. tRNA and protein were visualized on the gel by autoradiography and by Commassie staining, respectively. The results show that tRNA remains protein-bound at molar ratios of TsaE to TsaB2D2 below 2:1, i.e. at conditions under which only the TsaB2D2E1 complex is formed (Figure 5C and D). tRNA is rapidly displaced from the complex as the TsaE-to-TsaB2D2 molar ratio approaches 2:1, at which point all TsaE is sequestered in the TsaB2D2E2 complex and all tRNA is unbound, with excess free TsaE appearing at higher concentrations (Figure 5C, lanes 11–14 on the Commassie-stained gel). These results indicate that tRNA and TsaE occupy the same site on TsaB2D2 or that their binding sites significantly overlap. Notably, the fact that the fraction of bound tRNA remains unchanged as the complex transitions from TsaB2D2 to TsaB2D2E1 suggests that while the 2-fold structural symmetry of the TsaB2D2 complex, and the presence of two TsaE binding sites on it (Figure 4), are consistent with two tRNA binding sites on TsaB2D2, the complex can apparently accommodate only a single tRNA.
The observation that the T. maritima TsaB2D2E2 complex does not bind tRNA is consistent with previous reports of the orthologous, ternary complex from E. coli (E. coli TsaB1D1E1) failing to bind tRNA (23). Thus, we purified the E. coli binary complex TsaB1D1 and the ternary complex TsaB1D1E1 by gel filtration chromatography and investigated their tRNA binding properties. As in the T. maritima system, E. coli TsaB1D1 binds tRNA while the TsaB1D1E1 complex does not (Supplementary Figure S8), indicating conserved modes of binding of the TsaE subunit and of tRNA, and suggesting a conserved role of these interactions in the catalytic mechanism.
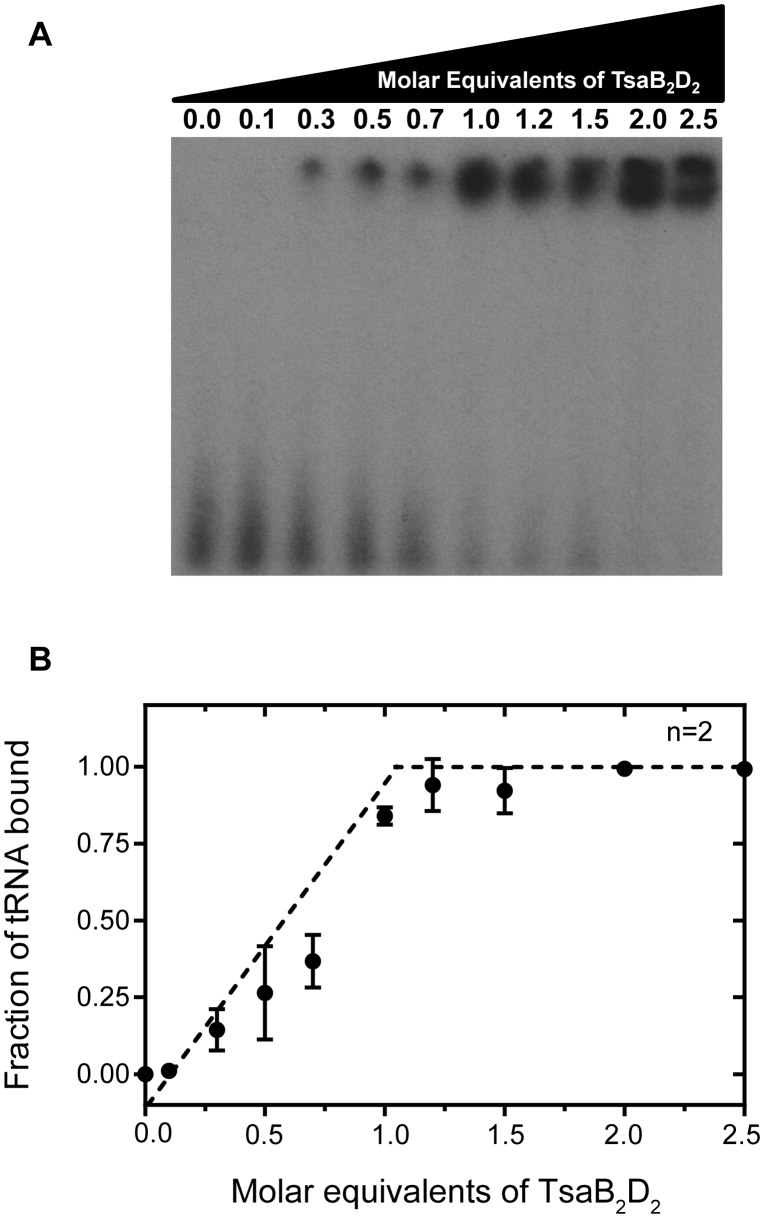
A single tRNA molecule binds in one of two equivalent sites on the TsaB2D2 platform
Given the results of the competitive EMSA experiment (Figure 5C and D), we conducted a gel mobility shift experiment using conditions where the tRNA concentration was ∼20-fold greater than the Kd in order to unambiguously determine the stoichiometry of tRNA binding to TsaB2D2. In this experiment, the pre-isolated TsaB2D2 complex was titrated into 32P-labeled tRNAThr, and the molar equivalent of protein necessary to saturate binding was determined by plotting the fraction of bound radiolabeled tRNA as a function of molar equivalents of protein to tRNA. The results clearly show saturation occurring at a tRNA-to-TsaB2D2 molar ratio of 1:1 (Figure 6), indicating that although two equivalent sites are available for tRNA binding on TsaB2D2, only one site can be filled at a time, indicating half-site reactivity.
Figure 6.
Stoichiometry of tRNA binding to TsaB2D2. (A) EMSA gel showing band shift corresponding to the TsaB2D2/tRNA complex with increasing molar equivalents of TsaB2D2. (B) Plot of fraction of tRNA bound quantified as the ratio of upper-band density to total density in each lane, as a function of the molar equivalents of the protein incubated with tRNA.
TsaB2D2 alone can catalyze a single turnover of t6A biosynthesis
The ability of tRNA to bind to TsaB2D2 as well as TsaB2D2E1, the fact that TsaD harbors the active-site responsible for the conversion of TC-AMP and tRNA to t6A-modified tRNA, and the observation that t6A activity assays in the absence of TsaE consistently exhibited higher DPM’s than those lacking any of the other three proteins (Supplementary Figure S2B), prompted us to investigate more closely the role and necessity of TsaE in t6A formation.
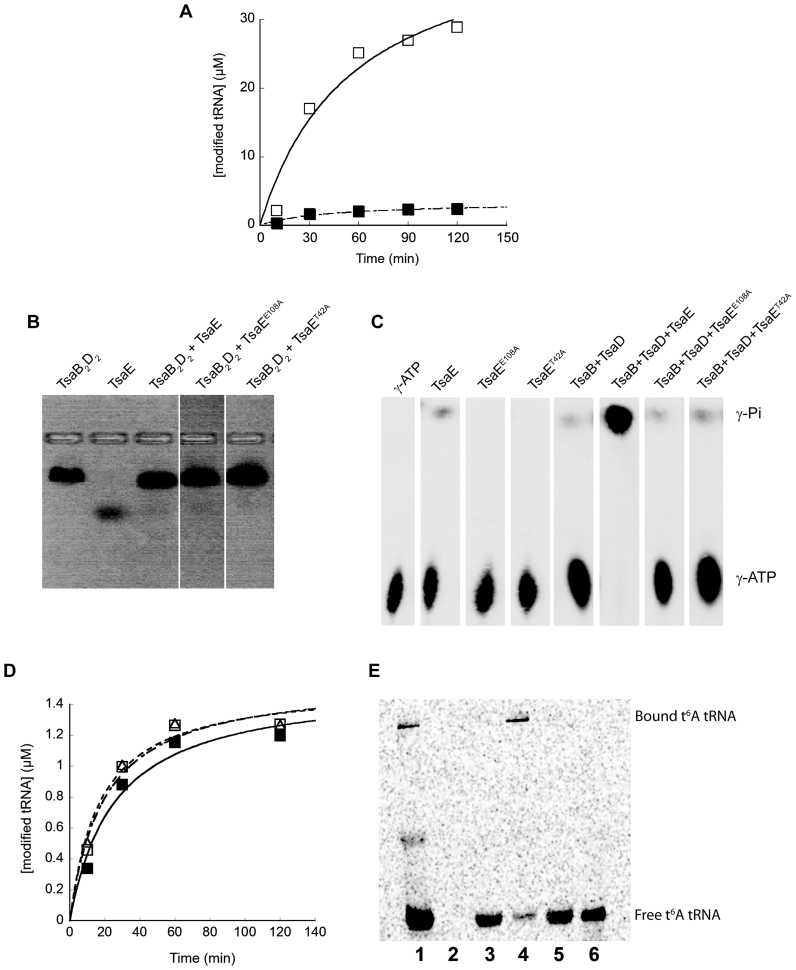
Inspection of our t6A assay data suggested that the absence of TsaE might result in a low level of activity, therefore we carried out t6A assays in the presence and absence of TsaE at higher substrate concentrations and longer times to better detect low levels of activity. The results (Figure 7A) show that while assays containing the TsaB2D2E1 complex result in robust activity with essentially complete conversion of substrate tRNA to t6A-modified tRNA, assays lacking TsaE produce t6A-modified tRNA up to but never exceeding the concentration of TsaB2D2, indicating that TsaB2D2 alone can only catalyze a single turnover. The rate of t6A formation by TsaB2D2 is also noticeably slower, with an initial velocity ∼3.5× lower than TsaB2D2E1-catalyzed t6A formation.
Figure 7.
TsaE is required for multiple turnover of the t6A biosynthesis reaction. (A) Standard radiochemical t6A assays measuring the incorporation of [14C]-threonine into tRNA. Assays contained Thermotoga maritima TsaC2 together with TsaB, TsaD and TsaE (open squares) or TsaB and TsaD (filled squares). (B) Native agarose gel-shift analysis showing the association of wild-type TsaE and mutants TsaEE108A and TsaET42A with TsaB2D2 in the absence of nucleotides. (C) TLC radiogram of the ATP hydrolysis reactions (generating ADP and Pi) catalyzed by wild-type TsaE, and mutants TsaEE108A and TsaET42A, in the absence and presence of TsaB2D2. [γ-32P] ATP and the radiolabeled hydrolysis product γ-phosphate are indicated. (D) Standard radiochemical t6A assays measuring the incorporation of [14C]-threonine into tRNA. Assays contained T. maritima TsaC2, TsaB and TsaD either alone (filled squares), or with added TsaEE108A (open squares) or TsaET42A (open triangles). (E) Radiochemical t6A assays ([14C]threonine) analyzed by native PAGE. Reaction assays were carried out as described in the ‘Materials and Methods’ section, and contained 8 μM each of TsaC2, TsaB and TsaD ([TsaB2D2] therefore 4 μM). The [tRNA] in assays analyzed in lanes 1, 3 and 5 was 40 μM, while the [tRNA] in the assay analyzed in lane 4 was 3.75 μM. Lane 1. Standard assay (assay contains TsaB, TsaD and TsaE). Lane 2. Control lane—assay lacking tRNA. Lane 3. Assay contains TsaB and TsaD. Lane 4. Assay contains TsaB and TsaD (low tRNA). Lane 5. Assay contains TsaB, TsaD and TsaEE108A. Lane 6. Control lane—[14C]-labeled t6A-modified tRNA.
The ATPase activity of TsaE is necessary for multiple turnovers
Given that TsaE exhibits robust ATPase activity (producing ADP and Pi) in the presence of TsaB and TsaD, we sought to determine whether this activity was coupled to the ability of TsaE to support multiple turnovers by TsaB2D2. To test this proposal, we constructed two active-site mutants of TsaE (TsaET42A and TsaEE108A) that are capable of binding to TsaB2D2 (Figure 7B and Supplementary Figure S9) but are deficient in ATP binding (Supplementary Figure S10) and hydrolysis (Figure 7C), and examined the ability of these mutants to support multiple turnovers in t6A assays. The results clearly show (Figure 7D) that the loss of ATPase activity in the mutant TsaE proteins is accompanied by an inability to support multiple turnovers in t6A enzyme activity assays.
To probe the timing of TsaE’s involvement in the reaction cycle, in particular whether it was required for the release of the t6A-modified product tRNA from TsaB2D2 or for a step after dissociation of product tRNA, we analyzed standard radiochemical activity assays via EMSA (Figure 7E). In these experiments the product t6A-modified tRNA becomes labeled with [14C]threonine, and can then be visualized after gel electrophoresis by standard phosphorimaging. As expected, in the assay containing all of the components of the TCT complex (Figure 7E, lane 1) there is a significant amount of t6A-modified tRNA produced, consistent with multiple turnovers, with most of the product tRNA migrating with the mobility of free tRNA (Figure 7E, lane 6) while a smaller amount of product tRNA can be observed bound to the TsaB2D2 (or TsaB2D2E1) complex (band near top of gel). When TsaE is absent (Figure 7E, lane 3), or a TsaE mutant deficient in ATP hydrolysis (E108A) is included in the assay (Figure 7E, lane 5), much less product is observed, consistent with the single turnover behavior revealed above (Figure 7A and D). Notably, however, all of the product tRNA runs as free tRNA, not as part of a TsaB2D2–tRNA complex, indicating that once t6A-modified tRNA is formed it does not remain bound to TsaB2D2, but freely dissociates from the TsaB2D2 complex. In these assays the [tRNA] is 10× that of [TsaB2D2], so that after a single turnover only a small fraction (≤0.1) of the total tRNA pool is [14C]-labeled t6A-modified product tRNA, and therefore virtually all tRNA bound to TsaB2D2 is unlabeled substrate tRNA. In contrast, when the [tRNA] is less than [TsaB2D2] (Figure 7E, lane 5), [14C]-labeled t6A-modified product tRNA represents the bulk of the total tRNA (as in lane 1) and partitioning of labeled tRNA between free and bound states is observed. In the absence of substrate tRNA (lane 2) there are no observable bands, confirming that all of the bands in the other lanes are due to t6A-modified tRNA and not to any non-specific association of [14C]threonine with assay components. A faint, diffuse band is also observed in lane 1 of Figure 7E between the band due to free tRNA and the TsaB2D2–tRNA complex, and may represent binding of tRNA to TsaC2 as observed above (Supplemental Figure S7).
Finally, the observation that TsaE binds to TsaB2D2 with higher affinity in the presence of ATP than in the absence of ATP (or the presence of ADP) suggested that t6A biosynthesis might involve the cycling of TsaE between a bound and unbound state with respect to TsaB2D2. To investigate this, we carried out t6A assays at variable [TsaE] to determine the TsaE-concentration dependence of the rate of t6A formation. The velocity data show a direct dependence on [TsaE] (Supplementary Figure S11) over a 10-fold concentration range, consistent with a mechanism in which TsaE binds and dissociates during each catalytic cycle.
DISCUSSION
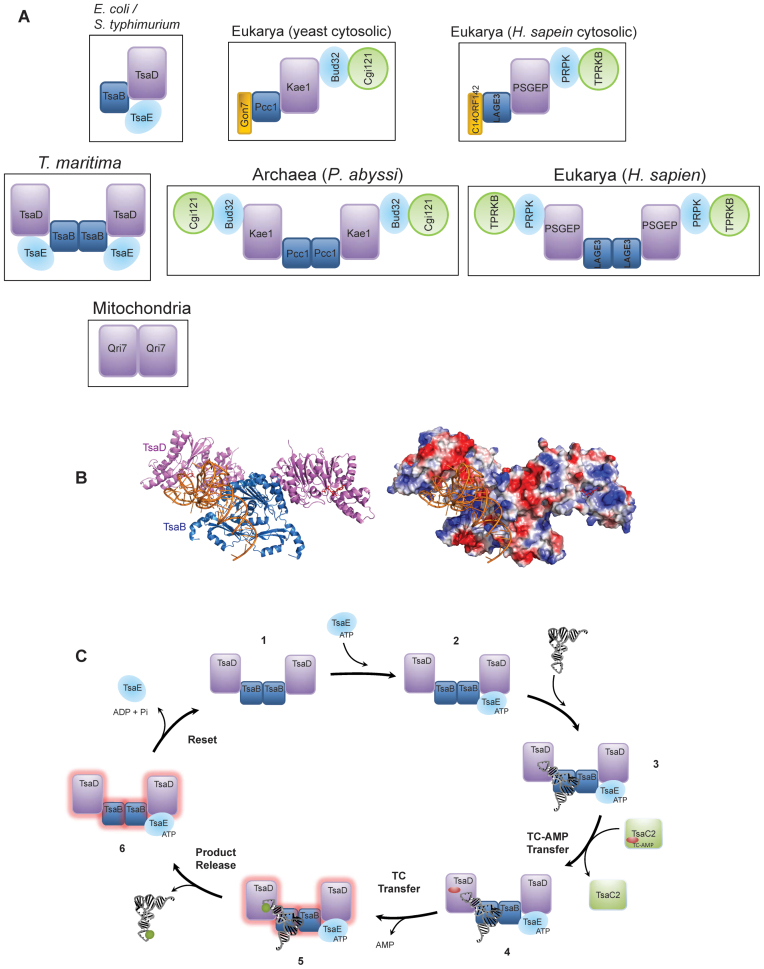
Species-specific architecture of the threonylcarbamoyl transfer (TCT) complex
The architecture of T. maritima TsaB2D2E2 complex presented here highlights the remarkable diversity in the assembly and quaternary architecture of the TCT complex in various species (Figure 8A). Comparing among bacteria, in E. coli, this complex exhibits the architecture of a single heterotrimer TsaB1D1E1 (23), whereas in T. maritima it is a dimer of heterotrimers. Similarly, the functionally analogous eukaryotic KEOPS complex is represented by a linear Pcc1-Kae1-Bud32-Cgi121 heterotetramer in yeast (capped on the Pcc1 side by the small subunit Gon7) (18), whereas the archaeal KEOPS complex is a dimer of Pcc1-Kae1-Bud32-Cgi121 heterotetramers (48). Notably, the orthologous KEOPS complex in human is the LAGE3-OSGEP-PRPK-TPRKB heterotetramer when capped on the LAGE3 side by the Gon7 ortholog C14ORF142, and a dimer of heterotetramers in the absence of C14ORF142 (17).
Figure 8.
Architecture of the TCT complex and role of its subunits in tRNA recognition and catalysis. (A) Schematic comparison of the architectures of the TCT complexes in Escherichia coli, Thermotoga maritima, Eukarya (yeast and human) and Archaea. The mitochondrial complex represented by a minimal Qri7 homodimer is also shown. The orthologous subunits in the various complexes are shown in the same color. (B) Ribbon diagram (left) and electrostatic surface representation (right) of the free-docking model of tRNA onto the T. maritima TsaB2D2 complex showing the positively charged, putative tRNA binding groove at the TsaB/D interface. Protein subunits are colored as in panel A. (C) Proposed catalytic cycle of t6A biosynthesis in T. maritima. Numbers represent the various complexes formed in the course of synthesis. Protein subunits are colored as in panel A. The red oval and green circle represent TC-AMP and t6A, respectively. The cycle is shown beginning with the TsaB2D2 structure (1). TsaE binds to one half of the preformed TsaB2D2 complex (2), while tRNA and TC-AMP bind to the other half (3 and 4) where transfer of TC to tRNA is catalyzed (5), releasing AMP and setting the system in an inactive state (glow). Following release of product tRNA (6), ATP hydrolysis by TsaE resets the system to enter the next cycle.
The larger assembly of the T. maritima TCT complex is attributed to the unique homodimerization mode of the TsaB subunit from this organism. Specifically, TsaB homodimerizes in two forms (46); form 1, common to all bacterial t6A systems studied to date (e.g. S. typhimurium (45), E. coli (49), V. parahaemolyticus (46) and T. maritima (41)), exhibits a hydrophobic helical interface and one of its monomers is replaced by the TsaD subunit in the complexes of T. maritima (Figures 3 and 4) and in the S. typhimurium and E. coli TsaB/TsaD complexes (23,24), i.e. the intersubunit interface in the heterocomplex mimics that in the form-1 TsaB homodimer. The form-2 TsaB homodimer, observed so far only in T. maritima (41), exhibits a different architecture in which the β-strands from the two TsaB subunits pack together in an anti-parallel manner to form an extended β-sheet, leaving the hydrophobic helical surface to interact with TsaD (Figures 3 and 4). This form-2 TsaB homodimer serves as the dimerization module in T. maritima TsaB2D2 and TsaB2D2E2 complexes, resulting in a dimer of heterodimers and heterotrimers, respectively. In these structures, the TsaB homodimer mimics the Pcc1 homodimer in the KEOPS complex in acting as a dimerization module for the entire complex (Figure 8A) (16,48). Therefore, although the form-1 TsaB homodimer is thought to be the dominant form, our results show that the form-2 homodimer is equally biologically relevant, at least in Thermotogae.
Docking model of tRNA onto the TsaB2D2 complex
We showed that the TsaB2D2 complex is the minimal tRNA binding scaffold (Figure 5 and Supplementary Figure S7), suggesting that the interface between TsaB and TsaD subunits is required for tRNA binding. The results also suggest that TsaE and tRNA occupy the same or overlapping site(s) on TsaB2D2. Consistent with these results, free docking of tRNA onto the SAXS structural model of the TsaB2D2 complex using HADDOCK (50) resulted in a model in which tRNA bound to a positively charged groove at the interface between TsaB and TsaD, which in turn places the anticodon loop of tRNA in the active site of the TsaD subunit (Figure 8B). This docking model shows tRNA occupying the TsaE binding site, consistent with the mutually exclusive binding of tRNA and TsaE observed by EMSA (Figure 5C and D). Further, attempts to dock a second tRNA molecule into the other TsaD subunit of TsaB2D2 resulted in severe clashes with the acceptor stem of the first tRNA (data not shown), consistent with a TsaB2D2-to-tRNA binding stoichiometry of 1:1 as observed by EMSA.
Proposed model of the catalytic cycle of t6A biosynthesis in bacteria
The ability of TsaB2D2 together with TsaC2 to catalyze a single cycle of t6A synthesis (Figure 7A) indicates that neither the TsaE subunit itself nor its ATPase activity is required for the transfer of TC-AMP from the TsaC2 active site to the TsaD active site, nor for the condensation of TC-AMP with tRNA to form t6A. Furthermore, the observation that product t6A-modified tRNA dissociates freely from the TsaB2D2 complex (Figure 7E) demonstrates that TsaE is also not required for tRNA release. Thus, the role of TsaE can be isolated to the post-reaction period, and suggests that the TsaB2D2 complex is left in a catalytically inactive state after formation of t6A-modified tRNA, and that TsaE-mediated ATP hydrolysis is required to somehow ‘reset’ the complex for another round of catalysis.
It has been reported that a mutant E. coli TsaE with no ATPase activity is still active in E. coli t6A biosynthesis (23). Similarly, it was reported that a mixture of B. subtilis TsaB, TsaD and TsaE can support the synthesis of t6A when provided with the isolated TC-AMP intermediate in the absence of ATP (10). These reports appear to be inconsistent with the data provided here for the T. maritima system, and may represent mechanistic differences in different bacterial species. However, the lack of rigorous t6A quantification in these cases combined with very low apparent turnover precludes us from determining the stoichiometry of product formation, and given that the protein concentrations used were reasonably high (2.5 and 5 μM), it is possible that the t6A produced in these studies represents a single turnover or less, and the requirement for TsaE-catalyzed ATP hydrolysis a mechanistic feature conserved in all bacterial systems; clarification of this issue awaits more detailed analysis of the reaction cycles for these systems.
The mutually exclusive binding of TsaE and tRNA, and the differential binding affinities of TsaE for TsaB2D2 (and TsaB1D1 in E. coli (23)) in the presence and absence of ATP, suggest that TsaE associates and dissociates in the course of a single catalytic cycle. Furthermore, although two TC transfer sites (TsaD) are present in TsaB2D2, only one product tRNA is turned over by this complex, indicating that TC transfer in one site renders the entire TsaB2D2 complex, not just one half of it, inactivated. This suggests that TsaE-catalyzed ATP hydrolysis in one half resets the entire TsaB2D2 platform, and that there is communication between the two halves of the complex during the course of t6A synthesis. Based on the data, we propose the following model for the t6A catalytic cycle in thermotoga. First, a single TsaE subunit (pre-bound to ATP) binds to the preformed heterocomplex TsaB2D2 (Figure 8C, 1 and 2), followed by tRNA binding (Figure 8C, 3). TsaC2 then binds, either with preformed TC-AMP or prior to TC-AMP synthesis, and delivers TC-AMP to the active site of the tRNA-bound TsaD subunit (Figure 8C, 4), allowing transfer of TC to tRNA (Figure 8C, 5). Following the formation of t6A, the t6A-modified tRNA dissociates from the complex (Figure 8C, 6). Subsequent hydrolysis of the bound ATP resets the system to the initial state, presumably by affecting a conformational change within the TsaB2D2 platform that occurred as a consequence of t6A formation, followed by release of TsaE. The next cycle of t6A synthesis can commence with the binding of TsaE bound to ATP.
There are several questions that remain unanswered by the data presented here and by our proposed mechanism. The observation that the initial velocity of t6A formation is greater (∼3.5-fold) in the assays containing TsaE than those without (Figure 7A) is surprising in light of the role of TsaE after product release (Figure 7E). This may be related to the apparent stabilizing effect of TsaE as seen in the TsaB2D2E2 complex (Figure 4) compared with the TsaB2D2 complex (Figure 3), coupled with the fact that in the absence of TsaE, TsaB2D2 functions as a stoichiometric reagent, not a catalyst; i.e. it is capable of only a single turnover and so can be considered ‘inactivated’ after it undergoes a reaction and thus is not available to catalyze another reaction. However, it may also be related to the interaction of TsaE with TsaC2 observed by native agarose (Figure 2), which may foreshadow another, albeit nonessential, role for TsaE in catalysis, distinct from the essential role associated with its ATPase activity.
Related to the nuances of the role of TsaE is the question of how TsaC2 interacts with the TCT complex, the timing of TC-AMP synthesis and transfer, and how the timing of TC transfer and TsaE-catalyzed ATP hydrolysis is controlled. TsaC2 clearly interacts with several components of the TCT complex (Figure 2A), as well as with tRNA (Supplemental Figure S7), but these interactions do not constitute the strong interactions observable, for example, by gel filtration chromatography, suggesting that they are transient. The details of these interactions, and their importance to the catalytic cycle, remain unknown at this point.
At last, and perhaps most intriguing, is the nature of the interactions between TsaB2D2 and TsaE associated with the ATPase activity of TsaE. It was unexpected that TsaE would function after product release, as opposed to in some way facilitating TC transfer or tRNA turnover. The observation that it instead appears to induce structural changes in TsaB2D2 is fascinating, and elucidating the full extent of these interactions clearly is important to understanding this modification process.
These questions and others are the subject of active investigation, the results of which will be reported in due course.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Robert Dowell for providing invaluable, critical feedback on the manuscript, Mr Rick McClintock for assistance with set up of the FPLC equipment, Dr Susan Krueger for assistance with the NMA and Dr Thomas Weiss for help with the SAXS data collection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM110588 to M.A.S., D.I.-R.]; California Metabolic Research Foundation; Western University of Health Sciences; U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences [DE-AC02–76SF00515]; Department of Energy Office of Biological and Environmental Research; National Institutes of Health; National Institute of General Medical Sciences [P41GM103393]. Funding for open access charge: National Institutes of Health; San Diego State University.
DISCLAIMER
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or the National Institute of General Medical Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1. Murphy F.V.T., Ramakrishnan V., Malkiewicz A., Agris P.F.. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 2004; 11:1186–1191. [DOI] [PubMed] [Google Scholar]
- 2. Agris P.F. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008; 9:629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. El Yacoubi B., Bailly M., de Crecy-Lagard V.. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012; 46:69–95. [DOI] [PubMed] [Google Scholar]
- 4. Grosjean H., de Crecy-Lagard V., Marck C.. Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett. 2010; 584:252–264. [DOI] [PubMed] [Google Scholar]
- 5. El Yacoubi B., Hatin I., Deutsch C., Kahveci T., Rousset J.P., Iwata-Reuyl D., Murzin A.G., de Crecy-Lagard V.. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J. 2011; 30:882–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Edvardson S., Prunetti L., Arraf A., Haas D., Bacusmo J.M., Hu J.F., Ta-Shma A., Dedon P.C., de Crecy-Lagard V., Elpeleg O.. tRNA N6-adenosine threonylcarbamoyltransferase defect due to KAE1/TCS3 (OSGEP) mutation manifest by neurodegeneration and renal tubulopathy. Eur. J. Hum. Genet. 2017; 25:545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Braun D.A., Rao J., Mollet G., Schapiro D., Daugeron M.C., Tan W., Gribouval O., Boyer O., Revy P., Jobst-Schwan T. et al. Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat. Genet. 2017; 49:1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thiaville P.C., Iwata-Reuyl D., de Crecy-Lagard V.. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t6A), a universal modification of tRNA. RNA Biol. 2014; 11:1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. El Yacoubi B., Lyons B., Cruz Y., Reddy R., Nordin B., Agnelli F., Williamson J.R., Schimmel P.R., Swairjo M.A., de Crecy-Lagard V.. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 2009; 37:2894–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lauhon C.T. Mechanism of N6-threonylcarbamoyladenonsine (t6A) biosynthesis: isolation and characterization of the intermediate threonylcarbamoyl-AMP. Biochemistry. 2012; 51:8950–8963. [DOI] [PubMed] [Google Scholar]
- 11. Deutsch C., El Yacoubi B., de Crécy-Lagard V., Iwata-Reuyl D.. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J. Biol. Chem. 2012; 287:13666–13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perrochia L., Guetta D., Hecker A., Forterre P., Basta T.. Functional assignment of KEOPS/EKC complex subunits in the biosynthesis of the universal t6A tRNA modification. Nucleic Acids Res. 2013; 4:9484–9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Srinivasan M., Mehta P., Yu Y., Prugar E., Koonin E.V., Karzai A.W., Sternglanz R.. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J. 2011; 30:873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Daugeron M.-C., Lenstra T.L., Frizzarin M., El Yacoubi B., Liu X., Baudin-Baillieu A., Lijnzaad P., Decourty L., Saveanu C., Jacquier A. et al. Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Res. 2011; 39:6148–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wan L.C., Mao D.Y., Neculai D., Strecker J., Chiovitti D., Kurinov I., Poda G., Thevakumaran N., Yuan F., Szilard R.K. et al. Reconstitution and characterization of eukaryotic N6-threonylcarbamoylation of tRNA using a minimal enzyme system. Nucleic Acids Res. 2013; 41:6332–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mao D.Y.L., Neculai D., Downey M., Orlicky S., Haffani Y.Z., Ceccarelli D.F., Ho J.S., Szilard R.K., Zhang W., Ho C.S. et al. Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine. Mol. Cell. 2008; 32:259–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wan L.C., Maisonneuve P., Szilard R.K., Lambert J.P., Ng T.F., Manczyk N., Huang H., Laister R., Caudy A.A., Gingras A.C. et al. Proteomic analysis of the human KEOPS complex identifies C14ORF142 as a core subunit homologous to yeast Gon7. Nucleic Acids Res. 2017; 45:805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang W., Collinet B., Graille M., Daugeron M.C., Lazar N., Libri D., Durand D., van Tilbeurgh H.. Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex. Nucleic Acids Res. 2015; 43:3358–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bacusmo J.M., Orsini S.S., Hu J., DeMott M., Thiaville P.C., Elfarash A., Paulines M.J., Rojas-Benitez D., Meineke B., Deutsch C. et al. The t6A modification acts as a positive determinant for the anticodon nuclease PrrC, and is distinctively nonessential in Streptococcus mutans. RNA Biol. 2017; 14:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Msadek T. Grasping at shadows: revealing the elusive nature of essential genes. J. Bacteriol. 2009; 191:4701–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Handford J.I., Ize B.R.R., Buchanan G., Butland G.P., Greenblatt J., Emili A., Palmer T.. Conserved network of proteins essential for bacterial viability. J. Bacteriol. 2009; 191:4732–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Butland G., Peregrin-Alvarez J.M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N. et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005; 433:531–537. [DOI] [PubMed] [Google Scholar]
- 23. Zhang W., Collinet B., Perrochia L., Durand D., van Tilbeurgh H.. The ATP-mediated formation of the YgjD–YeaZ–YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli. Nucleic Acids Res. 2015; 43:1804–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nichols C.E., Lamb H.K., Thompson P., Omari K.E., Lockyer M., Charles I., Hawkins A.R., Stammers D.K.. Crystal structure of the dimer of two essential Salmonella typhimurium proteins, YgjD & YeaZ and calorimetric evidence for the formation of a ternary YgjD–YeaZ–YjeE complex. Protein Sci. 2013; 22:628–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karst J.C., Foucher A.E., Campbell T.L., Di Guilmi A.M., Stroebel D., Mangat C.S., Brown E.D., Jault J.M.. The ATPase activity of an ‘essential’ Bacillus subtilis enzyme, YdiB, is required for its cellular function and is modulated by oligomerization. Microbiology. 2009; 155:944–956. [DOI] [PubMed] [Google Scholar]
- 26. Lesley S.A., Kuhn P., Godzik A., Deacon A.M., Mathews I., Kreusch A., Spraggon G., Klock H.E., McMullan D., Shin T. et al. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smolsky I.L., Liu P., Niebuhr M., Ito K., Weiss T.M., Tsuruta H.. Biological small-angle X-ray scattering facility at the Stanford Synchrotron Radiation Laboratory. J. Appl. Crystallogr. 2007; 40:s453–s458. [Google Scholar]
- 28. Huang T.C., Toraya H., Blanton T.N., Wu Y.. X-ray powder diffraction analysis of silver behenate, a possible low-angle diffraction standard. J. Appl. Crystallogr. 1993; 26:180–184. [Google Scholar]
- 29. McPhillips T.M., McPhillips S.E., Chiu H.-J., Cohen A.E., Deacon A.M., Ellis P.J., Garman E., Gonzalez A., Sauter N.K., Phizackerley R.P. et al. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiation. 2002; 9:401–406. [DOI] [PubMed] [Google Scholar]
- 30. Martel A., Liu P., Weiss T.M., Niebuhr M., Tsuruta H.. An integrated high-throughput data acquisition system for biological solution X-ray scattering studies. Journal of Synchrotron Radiat. 2012; 19:431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M., Gorba C., Mertens H.D.T., Konarev P.V., Svergun D.I.. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012; 45:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I.. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003; 36:1277–1282. [Google Scholar]
- 33. Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992; 25:495–503. [Google Scholar]
- 34. Schneidman-Duhovny D., Hammel M., Sali A.. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010; 38:W540–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Panjkovich A., Svergun D.I.. Deciphering conformational transitions of proteins by small angle X-ray scattering and normal mode analysis. Phys. Chem. Chem. Phys. 2016; 18:5707–5719. [DOI] [PubMed] [Google Scholar]
- 36. Schneidman-Duhovny D., Hammel M., Tainer J.A., Sali A.. FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 2016; 44:W424–W429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999; 76:2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Volkov V.V., Svergun D.I.. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003; 36:860–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marti-Renom M.A., Stuart A.C., Fiser A., Sanchez R., Melo F., Sali A.. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000; 29:291–325. [DOI] [PubMed] [Google Scholar]
- 40. Sali A., Blundell T.L.. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993; 234:779–815. [DOI] [PubMed] [Google Scholar]
- 41. Xu Q., McMullan D., Jaroszewski L., Krishna S.S., Elsliger M.-A., Yeh A.P., Abdubek P., Astakhova T., Axelrod H.L., Carlton D. et al. Structure of an essential bacterial protein YeaZ (TM0874) from Thermotoga maritima at 2.5 Å resolution. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010; 66:1230–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pierce B.G., Wiehe K., Hwang H., Kim B.-H., Vreven T., Weng Z.. ZDOCK server: interactive docking prediction of protein–protein complexes and symmetric multimers. Bioinformatics. 2014; 30:1771–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ryder S.P., Recht M.I., Williamson J.R.. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol. Biol. 2008; 488:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nichols C.E., Johnson C., Lockyer M., Charles I.G., Lamb H.K., Hawkins A.R., Stammers D.K.. Structural characterization of Salmonella typhimurium YeaZ, an M22 O-sialoglycoprotein endopeptidase homolog. Proteins. 2006; 64:111–123. [DOI] [PubMed] [Google Scholar]
- 46. Aydin I., Saijo-Hamano Y., Namba K., Thomas C., Roujeinikova A.. Structural analysis of the essential resuscitation promoting factor YeaZ suggests a mechanism of nucleotide regulation through dimer reorganization. PLoS One. 2011; 6:e23245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vecchietti D., Ferrara S., Rusmini R., Macchi R., Milani M., Bertoni G.. Crystal structure of YeaZ from Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 2016; 470:460–465. [DOI] [PubMed] [Google Scholar]
- 48. Wan L.C., Pillon M.C., Thevakumaran N., Sun Y., Chakrabartty A., Guarne A., Kurinov I., Durocher D., Sicheri F.. Structural and functional characterization of KEOPS dimerization by Pcc1 and its role in t6A biosynthesis. Nucleic Acids Res. 2016; 44:6971–6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jeudy S., Stelter M., Coutard B., Kahn R., Abergel C.. Preliminary crystallographic analysis of the Escherichia coli YeaZ protein using the anomalous signal of a gadolinium derivative. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2005; 61:848–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Zundert G.C., Rodrigues J.P., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S., van Dijk M., de Vries S.J., Bonvin A.M.. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016; 428:720–725. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.