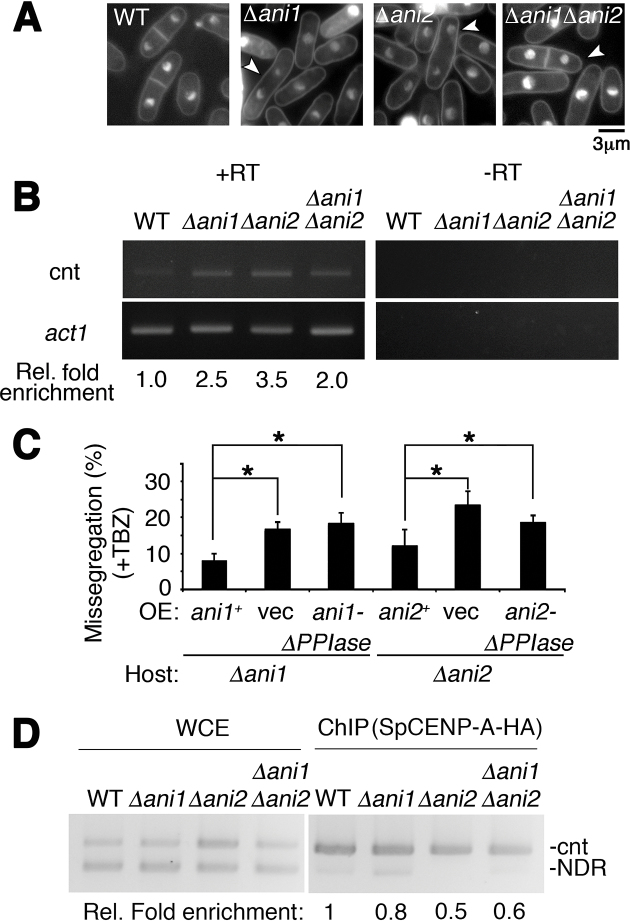
Figure 4.
Catalytic function of Ani1 and Ani2 is required for regulation of precise chromosome segregation. (A) Nuclear phenotypes of wild type (WT), Δani1, Δani2 and Δani1Δani2. Cells harboring Δani1, Δani2 and Δani1Δani2 all showed unequal chromosome segregation. White arrowhead indicates missegregated bi-nucleated cells. Bar: 3 μm. (B) Transcription de-repression of centromeric DNA following the concurrent disruption of ani1 and ani2. WT, wild type; Ani, SpCENP-A NTD-isomerase. +RT, –RT: with and without reverse transcription, respectively; cnt, central core region of centromere 1; Act1, actin normalizing control. Representative gel image of two biological replicates. Rel. fold enrichment: fold enrichment relative to WT. (C) Mutant ani1 and ani2 genes containing mutation in catalytic domains cannot suppress chromosome segregation defects of Δani1 and Δani2. Frequency of the chromosome missegregation phenotype of strains expressing vector only (vec), full-length ani1+ and ani2+, and isomerase domain null (-ΔPPIase) proteins in ani1 or ani2 deletion strains. Assay was performed 18 h post-induction in EMM-leucine after thiamine wash-out at 26°C followed with 3 h treatment in thiabendazole (TBZ). Missegregation phenotypes were suppressed in Full-length but not in the isomerase domain null mutant. Vec, empty vector control. N = 150. Error: S.D. Mean of three biological replicates.Two tailed t-test. *P < 0.05. (D) ChIP assay for SpCENP-A-HA in WT, Δani1, Δani2 and Δani1Δani2 cells. cnt: SpCENP-A at the central core region of centromere 1. WCE, Whole cell extract; NDR, nucleosome-depleted region as normalization control. Representative image and average enrichment of two independent experimental replicates.