Abstract
Introduction:
Phylogenetic relationships between different lineages of Trypanosoma cruzi, the agent of Chagas disease, have been controversial for several years. However, recent phylogenetic and phylogenomic analyses clarified the nuclear relationships among such lineages. However, incongruence between nuclear and kinetoplast DNA phylogenies has emerged as a new challenge. This incongruence implies several events of mitochondrial introgression at evolutionary level. However, the mechanism that gave origin to introgressed lineages is unknown. Here, I will review and discuss how maxicircles of the kinetoplast were horizontally and vertically transferred between different lineages of T. cruzi.
Conclusion:
Finally, I will discuss what we know - and what we don't - about the kDNA transference and inheritance in the context of sexual reproduction in this parasite.
Keywords: Trypanosoma cruzi, DTU, Evolution, Phylogeny, Hybridization, Kinetoplast, Mitochondrial introgression
1. INtroduction
1.1. Kinetoplatids and Sexual Reproduction
Kinetoplastids are a group of unicellular heterotrophic eukaryotes including several species with medical or ecological importance. Although this group has some unorthodox solutions to common problems of eukaryotic cells, the main common characteristic is the single mitochondrion with its DNA organized in a really complex network [1-3]. This network is known by the term kinetoplast and its DNA as kDNA. Kinetoplastids cluster together with diplonemids in the subphylum Glycomonada of the phylum Euglenozoa [4], one of the candidates as the most ancient branch in the eukaryotic tree of life [5, 6]. Although it was assumed for several years that such clades were asexual, it is currently known that sexual reproduction (meiosis + mating) is as ancient as the eukaryotes [7, 8]. However, although sex may occur, it does not always happen. Even some species of kinetoplastids may have lost the ability of sexual reproduction [9]. However, in most of the species, it has been proposed that sex is not an obligate step for the organism and only occurs in some situations [8]. In this regard, Tibayrenc and Ayala have proposed that parasitic kinetoplastids have a predominant clonal evolution [10, 11]. They argued that genetic exchange is restrained at the population level, or at least, mainly occurs between genetically identical organisms (selfing or inbreeding). This model was challenged by several authors and controversy is still installed [12-15]. Despite the debate on the true impact of sexual exchange to the population structure of different kinetoplastids, there are several evidences that sex may occur (because it was observed in the laboratory) [16, 17] or it has already happened (because of its traces in phylogenetic and population genetic analyses) [18, 19].
2. SEXUAL EXCHANGE IN TRYPANOSOMA
Most of the evidences of genetic exchange have been shown for trypanosomes. Around ninety species described in the genus Trypanosoma infect a wide range of vertebrates and they are transmitted by blood-sucking arthropods (insects and ticks) and leeches. Few species are implicated in human disease. Although atypical infections in humans caused by T. vivax, T. congolense, T. evansi, T. lewisi, and T. lewisi-like have been reported [20], T. brucei and T. cruzi are the most common causes of human trypanosomiasis (sleeping sickness and Chagas disease respectively). In this regard, because the genetic exchange is a common way to disperse virulence factors or any other undesirable medical characteristics, the sexual exchange in such trypanosomes was actively focused on in the last decades. Particularly, evidence of the formation of haploid gametes and mating has been shown for T. brucei and several papers had success to get recombinants (reviewed in [16]). However, the finding of sexual reproduction in T. cruzi was elusive. Only one paper described the formation of a genetic hybrid [21]. However, meiosis and gametes have not been observed yet. Despite, there are several phylogenetic and population genetic evidences of events of genetic exchange [22-28] and even two lineages show characteristics of a meiotic F1 [29]. However, a different phenomenon of hybridization in T. cruzi, the mitochondrial introgression [22, 25, 30, 31] was reported. This phenomenon consists of the observation by genetic analysis of a particular hybrid which has the mitochondrial genome (kDNA in this case) of one lineage but a nuclear background from a different one. Mechanisms for the formation of such hybrids are unknown in Trypanosoma. Below we describe and discuss the evidence of such hybrids and how the kDNA was transferred between different lineages.
3. TRYPANOSOMA CRUZI KDNA: A BRIEF DESCRIPTION OF THE TRAVELLER PROFILE
The kDNA structure is composed of several thousands of circular DNA molecules which are concatenated in a complex network (see [32] for a detailed review kDNA in kinetoplastida). There are two different DNA types in this network: maxicircles and minicircles. There are a few dozen of maxicircles in the kDNA and they code for different mitochondrial proteins. In T. cruzi, the maxicircles have 20 genes [33]. Particularly, nine out of them are cryptogenes because their DNA sequences are very different from their mature mRNA. Such cryptogenes are transcribed and the immature mRNA needs a complex system of edition in order to be a fully functional transcript (reviewed in [34]). The edition of mRNA is guided by short RNAs called guide RNA or gRNA and most of such gRNA are coded by minicircles. There are 20,000-30,000 minicircles in the kinetoplast of T. cruzi and each minicircle has four constant regions flanked by hyper-variable sequences [35]. The last ones code for gRNAs. Consequently, there are several thousands of different gRNA in a single parasite. Such gRNA can be sorted in different classes according to the sequence they edit. If a class of minicircles is lost, the target mRNA cannot be edited and thus a functional protein cannot be synthetized. Consequently, it is important to correctly duplicate and segregate minicircles during the kDNA division. Basically, each minicircle is released from the network, then it is replicated and finally both are linked to new networks in antipodal sites [36]. However, the system may fail to correctly distribute minicircles between both new kDNAs [37]. Consequently, minicircles and maxicircles are subjected to a certain degree of genetic drift. Although replication and segregation of the kDNA have been broadly studied, the behaviour of such complex network in sexual reproduction is mainly unknown. In T. brucei hybrids, kDNA is bi-parentally inherited which probably implies fusion of the parental mitochondria [38]. However, genetic drift homogenises maxicircle sequences in few generations [38]. Consequently, inheritance is just apparently uniparental for maxicircles. In T. cruzi, maxicircles from hybrid DTUs TcV and TcVI are similar to maxicircles from the parental TcIII [31, 33, 39, 40]. However, it was not addressed whether the maxicircles of TcII parental were not inherited or they were lost by homogenization as observed in T. brucei. In addition, minicircle inheritance was not addressed yet.
4. NUCLEUS AND KINETOPLAST DO NOT TRAVEL TOGETHER PART I: THE NUCLEAR JOURNEY
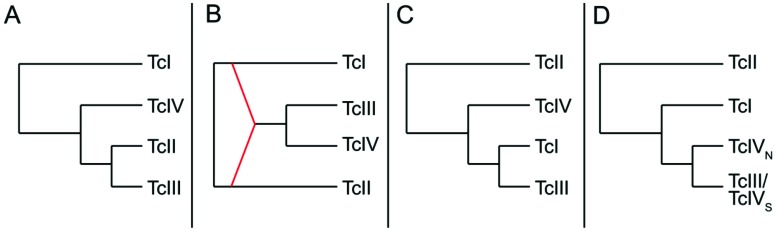
Currently, T. cruzi is divided into six discrete typing units (DTUs) called TcI to TcVI [41, 42]. The phylogenetic relationships among them were extensively studied. There is strong evidence supporting that two DTUs (TcV and TcVI) have their origin in nuclear hybrids between TcII and TcIII [31, 39, 43, 44]. Additional evidences of genetic exchange in T. cruzi have been inferred by detecting incongruence in the phylogeny for different genes or markers. When two genes have different evolutionary stories, it means that genetic exchange was implied. Consequently, phylogenetic incongruence between different genes or genomic regions is an indicium of genetic exchange. If incongruence is detected between nuclear and mitochondrial genes but not between different nuclear genes, the term mitochondrial introgression is used. Consequently, confident phylogenies of nuclear and mitochondrial markers are required to get a confident evidence of mitochondrial introgression. The nuclear phylogeny of different DTUs was controversial for several years and different models for the relationships among different DTUs are shown in (Fig. 1).
Fig. (1).
Different models of nuclear or kDNA relationships among DTUs TcI, TcII, TcIII and TcIV. A) The model proposes that TcIII and TcIV clusters with TcII according nuclear markers; B) The model proposes that TcIII and TcIV are hybrids between TcI and TcII according nuclear markers; C) The model proposes that TcIII and TcIV clusters with TcI according nuclear markers; D) The model proposes that TcIII and TcIV clusters with TcI, and TcIII and TcIV are not monophyletic groups according kDNA markers.
Brisse and coworkers proposed the first model for the relationships of DTUs and the current division into six DTUs [45]. This first model was based on Multilocus Enzyme Electrophoresis (MLEE) and Random Amplified Polymorphic DNA (RAPD) for 49 stocks of T. cruzi. MLEE is based on electrophoretic patterns of enzymes codified by housekeeping genes whereas RAPD gets a random overview of the genome by using a PCR with short random primers. The model proposed two main clusters: TcI and TcII-TcIII-TcIV (this last group also included hybrids TcV and TcVI but they were not included here for simplicity). However, other papers that analysed sequence data questioned this model [39, 43, 46, 47]. In a previous paper, it was proposed that the clustering observed by Brisse and coworkers was biased by the inclusion of hybrid DTUs TcV and TcVI in the analysis [23]. A simple simulation of MLEE data was made based on the multi-locus sequences showing that tree topology by MLEE data is strongly modified by the inclusion of hybrids. Although the analysis of the simulated MLEE data without including the hybrids showed the cluster TcIII-TcI-TcIV, the inclusion of the hybrids biased the analysis and instead clustered TcIII with TcII.
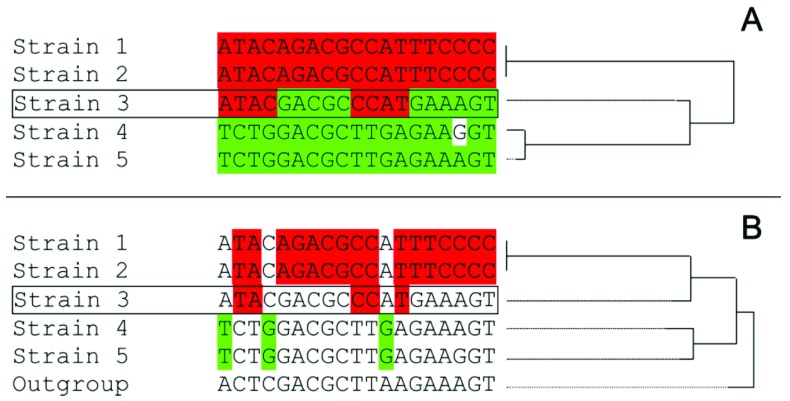
The second model about DTU relationships (Fig. 1B) proposed an ancient hybridization between TcI and TcII as the origin of TcIII and TcIV [46-48]. The observation of inconsistencies of sequence data against the model A supported this model. Initially, the hypothesis of the hybrid origin of TcIII and TcIV was based on the analysis of nine loci in representative strains of different DTUs [46, 47]. Six out of nine such loci corresponded to histones and heat shock proteins which have multiple copies in the genome. It was observed that certain SNPs of TcIII and TcIV were shared with TcII, whereas, others were shared with TcI. Consequently, the authors proposed that such sequences of TcIII and TcIV were mosaics of TcI and TcII. However, such mosaicism is only apparent because the authors did not include an outgroup in their analyses. In phylogenetic analysis, a character shared by a group of taxa is the only evidence of a common ancestor, if such character is not shared with an outgroup (synapomorphy). Instead, if the character is shared with the outgroup, it is probably an ancestral feature (plesiomorphy). Most of the SNPs that apparently clustered TcIII or TcIV with TcII were also shared with the outgroup (plesiomorphy) and consequently, they did not support such clustering (see [23]). Fig. (2) shows an example of apparent mosaicism that is solved by the inclusion of an outgroup. In addition, the authors proposed that four loci showed shorter distances between TcIII/TcIV and TcII [46]. However, despite the shorter distances, including an outgroup did not support the clustering of TcIII/TcIV with TcII [23].
Fig. (2).
Example of a false mosaic sequence caused by plesiomorphies. A) Alignment of sequences with a potential recombinant sequence (Strain 3). Note that the midpoint-rooted Neighbour Joining tree cluster such sequence with strains 4 and 5 (distance from strain 3 to strains 4 and 5 is shorter than distance to strains 1 and 2). B) The same alignment than in A but including an outgroup strain. Note that only sinapomorphies are highlighted and there is no SNP supporting the clustering of strain 3 with strains 4 and 5. Instead, there are five SNPs supporting the clustering of Strain 3 with strains 1 and 2. In addition, a rooted Neighbour Joining tree (right) show the clustering of Strain 3 with strains 1 and 2.
Recent papers also used more sophisticated methods of analysis of mosaicism such as BOOTSCAN [49] to demonstrate recombination [50, 51]. However, serious methodological concerns were observed. For example, Franco and co-workers [50] showed a mosaic sequence in the gene for the ABCG-like transporter for a TcIII strain with TcI and TcII as putative parentals. However, the authors used the “close relative” option in RDP software which is only recommended when putative parentals are more similar between them than with the recombinant (it is not the case here). In addition, they did not inform p values for each potential recombination event (bootstrap value is only for the detection of potential recombinant regions; a binomial test should be used to evaluate significance in order to avoid false positives according [49]). The analysis with their sequences in the same conditions used by the authors was repeated and no statistically significant events were detected using the binomial test with or without a Bonferroni correction. Similar concerns have also been detected [51]. Other papers that support the ancient hybridization between TcI and TcII were based on networks using the neighbour-net analysis and showing reticulate patterns [52, 53]. However, reticulate pattern in neighbour-net only shows character inconsistency which may be caused by different phenomena (i.e. homoplasy, paralogy, etc) instead of recombination [54]. In addition, supported splits of phylogenetic networks shown in such papers are fully compatible with the third model described below (See Fig. (2) in [53] supporting the TcI-TcIII cluster according to the 195 bp satellite sequences with high bootstrap support). Moreover, the analysis of the CL-Brener genome, a representative strain of the hybrid TcVI revealed very few sequences (less than 1% of the core genome) as candidates for mosaicism in the TcIII-derived counterpart of the genome [55]. Other papers presented data proposing that sequences similar to TcI in the genome of CL-Brener are the evidence of the ancient hybrid origin of TcIII [56, 57]. However, such data are not conclusive because they are also compatible with the third model that proposes TcI and TcIII are closely related. This relatedness between TcI and TcIII may explain the similarity between TcI and CL-Brener sequences.
The third model proposes that the T. cruzi ancestor diverged into two main groups (TcII and TcI-TcIII-TcIV). Posteriorly, TcI-TcIII-TcIV was divided into two groups TcI-TcIII and TcIV. Finally, TcIV diverged into TcIVS (from South America) and TcIVN (from North America) nearly at the same time that TcI and TcIII diverged into two different DTUs. The model is supported by the analysis of nuclear sequences from 13 single-copy housekeeping genes in 18 strains [23, 58]. In addition, the same phylogeny was also supported by analysing sequences for thirty-two protein coding regions in seven strains [43]. Other papers analysing few loci also give evidence of this model [31, 59].
Finally, although a single hybridization event was proposed for the origin of TcV and TcVI [40] as the most parsimonious hypothesis, more recent papers support two independent hybridization events [23, 31].
5. NUCLEUS AND KINETOPLAST DO NOT TRAVEL TOGETHER PART II: THE KDNA JOURNEY
Phylogenetic relationships of maxicircle sequences of different DTUs were not as controversial as the nuclear phylogeny. Basically, three main clades were observed (TcI, TcII and TcIII-TcIV) which are incongruent with nuclear clustering (Fig. 1D) and constitutes evidence of mitochondrial introgression. Machado and Ayala were the first to describe three different kDNA clades based on the sequence of two maxicircle genes (NADH dehydrogenase subunit 1 and Cytochrome Oxidase subunit II) [39]. The three kDNA clades were also observed with an additional maxicircle gene (Cytochrome b) and in a more extensive number of strains [40].
In addition, a phylogenomic analysis of the entire maxicircle sequences of Sylvio x10 (TcI), Esmeraldo (TcII), CL-brener (TcVI which inherited maxicircles only from TcIII) corroborated that clades TcI and TcIII-TcIV are joined in a major clade [33]. More recent papers showed that TcIII-TcIV may be divided into two main clades TcIVN and TcIVS-TcIII showing that both DTUs are not monophyletic for kDNA [23, 25, 31, 60].
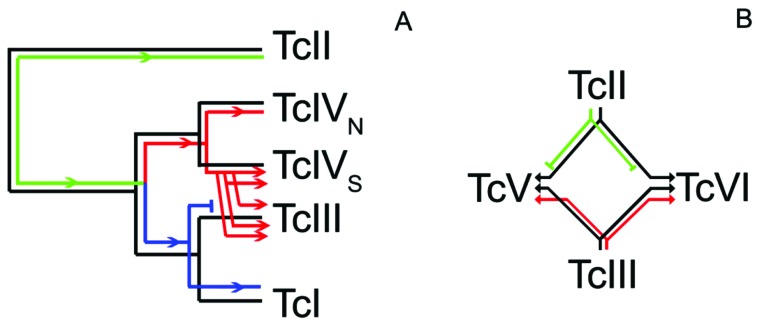
Joining nuclear and maxicircle phylogenies gives information about kDNA transfer among different DTUs (Fig. 3). An ancient separation between TcII and TcI-TcIII-TcIV is supported by both nuclear and maxicircle genes. Posteriorly, TcIV separated from TcI-TcIII according to the nuclear data. Finally, TcIV separated into TcIVN and TcIVS whereas TcI-TcIII diverged in the current DTUs. Incongruence between nuclear and maxicircle sequences may be explained by kDNA transfer from one DTU to another. Consequently, two alternative hypotheses are possible according to the direction of the kDNA transfer: from TcIII to TcIVs or from TcIVs to TcIII. Considering that TcI-TcIII and TcIVS-TcIVN are monophyletic clades, if TcIII transferred its kDNA to TcIVS it would be expected that TcIII-TcIVS clustered with TcI in the kDNA phylogeny. Instead, TcIII-TcIVS clusters with TcIVN suggested the alternative way: TcIVS transferred its kDNA to TcIII. In a recent paper it was proposed that such transference should have occurred several times in the history of TcIII (at least three times always from TcIVS to TcIII) [23]. Finally, TcIII transferred this TcIVs kDNA to hybrids TcV and TcVI in two independent hybridization events [23]. Finally, these evolutionary events of mitochondrial introgression are also supported by the observation of several recent events between TcIV and TcI [22, 25, 30].
Fig. (3).
kDNA (maxicircle) journey in the evolution of different DTUs of Trypanosoma cruzi. Nuclear (black lines) and maxicircle (coloured lines) phylogenies are overlapped. Different colours in kDNA phylogeny represent the three different major kDNA clades, and arrows represent how kDNA is transmitted. A) Nuclear and maxicircle phylogenies are mainly congruent with the exception of TcIII clustering with TcIVs (kDNA phylogeny) instead of TcI (nuclear phylogeny). This incongruence is explained by the transference of the kDNA from TcIVS to TcIII. In addition, note that TcIII and TcIVS are not monophyletic clades according to kDNA which may be explained by several events of introgression (at least three events, see [23]). Note that the ancestral TcIII kDNA was lost (interrupted blue line). B) Schematic network showing two independent TcII/TcIII hybridizations. Note that TcII and TcIII contribute with their nuclear genomes to hybrids but maxicircles are only from TcIII. Maxicircles from TcII (green lines) were not transferred or they were lost by drift after the hybridization.
6. DO WE KNOW THE MECHANISMS OF HYBRIDIZATION?
Gaunt and coworkers proposed a mechanism of genetic exchange in T. cruzi several years ago [21]. The model proposes the formation of a tetraploid hybrid and posterior chromosome loss to return to a state near the diploidy. This model was based on the observation of a tetraploid hybrid in mammal cell cultures infected with two different strains of T. cruzi. However, if a random loss of chromosomes (or genes) occurs to return diploidy, it is expected that some genes will have lost both copies of the same parental. Consequently, a homozygous state would be expected randomly in 1/3 of the genome [29]. However, this expectancy is far from the observed heterozygosis in the genomic data of CL-Brener strain and other strains from hybrid DTUs [58, 61]. These DTUs resemble a typical F1 after meiosis. Although meiosis and gametes were not observed for T. cruzi, both are not unlikely because they were observed in experimental crosses in the relative T. brucei. In addition, the genome of T. cruzi conserved the whole machinery for homologous recombination required in meiosis [44].
The mechanism of mitochondrial introgression is also unknown and several questions arise from the observation of such phenomenon. The first question is how kDNA is introgressed. There are two possible mechanisms for introgression: successive backcrosses or mitochondrial exchange. The first is the mechanism observed in superior organisms. As a hypothetical example of the introgression of kDNA from TcIVs to TcIII, the first step would be the occurrence of a meiotic hybrid TcIII/TcIVS. This hypothetical hybrid inherited the kDNA from TcIVS. Posteriorly, successive backcrosses with TcIII would have reduced the proportion of TcIVS nuclear genome on the hybrids although TcIVS kDNA was maintained. It is important to note that the proportion of TcIVS nuclear genome in the hybrid would be in average 0.5n, where n is the number of backcrosses. Consequently, only 10 backcrosses of the TcIII-TcIVS hybrids with TcIII will reduce the proportion of TcIVS genome to less than 0.1%. The main drawback of this hypothesis about the mechanism of introgression is that it requires relatively frequent events of meiotic sexual exchange in the past and this was not detected in T. cruzi. The alternative method, mitochondrial exchange is only hypothetical and it simply consists of the exchange of mitochondria or their kDNA between parasites. It is based on the observation mitochondrial transfer between mammal cells [62]. This phenomenon is able to rescue deleterious mitochondrial genotypes in some cells if they are surrounded by cells with normal genotypes. Here it is important to note that the mitochondrion of the T. cruzi is the candidate which suffers from the Muller’s ratchet because of the asexual mode of reproduction of this organelle [63]. The Muller’s ratchet hypothesis proposes that an asexual population will undoubtedly be extinct because of the accumulation of deleterious mutation (sexual reproduction allows escaping the ratchet). Although multiple copies of maxicircles and minicircles may help to avoid the Muller’s ratchet [64], it may not be enough and mitochondrial or kDNA exchange may help the parasite to avoid the ratchet.
The second question is related to the asymmetrical transference of the kDNA. Why the kDNA was transferred in the same direction several times (from TcIVS to TcIII) and why both hybrid DTUs TcV and TcVI inherited such kDNA in independent hybridizations? Is there an evolutionary advantage in the kDNA of TcIVS? In a previous paper it was proposed that such asymmetrical introggression from TcIVS to TcIII may also be explained by neutral demographic models (i.e. selective advantage is not implied). The model proposes that when a species invades an area already occupied by a related species, asymmetrical introgression may occur mainly from the local species towards the invader [65]. Such asymmetrical mitochondrial introgression was observed for several animal and plant species [65] and even in algae [66]. However, the major drawback of such a model is the requirement of frequent genetic exchange between TcIVS and TcIII at least in front of the expansion wave.
The third question about introgression is related to the inheritance of the kDNA, because biparental inheritance cannot be discarded. In this regard, although there is evidence that TcIII received maxicircle sequences from TcIVS, it is not clear if the whole minicircle sequences in TcIII also came from TcIVs.
CONCLUSION / FUTURE PERSPECTIVES
Mitochondrial introgression (at least maxicircle introgression) has occurred in the evolutionary history of T. cruzi. The transference of kDNA between different DTUs is shown in Fig. (3). However, the mechanism and biological importance of such transference are completely unknown. Understanding the inheritance of minicircles in the hybrid DTUs (is it uniparental or biparental?) will help to understand if mitochondrion fusion is possible. The main problem in the analysis of minicircle phylogeny is the high variability and the high number of copies which makes it difficult to address the question with conventional tools. However, next generation sequencing methods may help in order to get data about populations of minicircles in strains of different DTUs. Finally, looking for other mechanisms of nuclear genetic exchange than the previously observed in the laboratory is still relevant to understand the mechanism of introgression and to explain why TcV and TcVI are mainly heterozygous.
CONSENT FOR PUBLICATION
Not applicable.
ACKNOWLEDGEMENTS
I acknowledge to Dr. Patricio Diosque for useful discussions about evolution and phylogeny.
CONFLICT OF INTEREST
The author declares no conflict of interest. This work has been supported by PICT-2014-2449 ANPCyT.
REFERENCES
- 1.Moreira D., Lopez-Garcia P., Vickerman K. An updated view of kinetoplastid phylogeny using environmental sequences and a closer outgroup: Proposal for a new classification of the class Kinetoplastea. Int. J. Syst. Evol. Microbiol. 2004;54(Pt 5):1861–1875. doi: 10.1099/ijs.0.63081-0. [DOI] [PubMed] [Google Scholar]
- 2.Simpson A.G., Stevens J.R., Lukes J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;22(4):168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Stevens J.R. Kinetoplastid phylogenetics, with special reference to the evolution of parasitic trypanosomes. Parasite. 2008;15(3):226–232. doi: 10.1051/parasite/2008153226. [DOI] [PubMed] [Google Scholar]
- 4.Cavalier-Smith T. Higher classification and phylogeny of Euglenozoa. Eur. J. Protistol. 2016;56:250–276. doi: 10.1016/j.ejop.2016.09.003. https://www.sciencedirect.com/science/article/pii/S0932473916300839 [DOI] [PubMed] [Google Scholar]
- 5.Cavalier-Smith T. The neomuran revolution and phagotrophic origin of eukaryotes and cilia in the light of intracellular coevolution and a revised tree of life. Cold Spring Harb. Perspect. Biol. 2014;6(9):a016006. doi: 10.1101/cshperspect.a016006. http://cshperspectives. cshlp.org/content/6/9/a016006.full.pdf+html [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raymann K., Brochier-Armanet C., Gribaldo S. The two-domain tree of life is linked to a new root for the Archaea. Proc. Natl. Acad. Sci. USA. 2015;112(21):6670–6675. doi: 10.1073/pnas.1420858112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodenough U., Heitman J. Origins of eukaryotic sexual reproduction. 2014 doi: 10.1101/cshperspect.a016154. http://cshperspectives.cshlp.org/content/6/3/a016 [DOI] [PMC free article] [PubMed]
- 8.Speijer D., Lukes J., Elias M. Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life. Proc. Natl. Acad. Sci. USA. 2015;112(29):8827–8834. doi: 10.1073/pnas.1501725112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weir W., Capewell P., Foth B., Clucas C., Pountain A., Steketee P., Veitch N., Koffi M., De Meeus T., Kabore J., Camara M., Cooper A., Tait A., Jamonneau V., Bucheton B., Berriman M., MacLeod A. Population genomics reveals the origin and asexual evolution of human infective trypanosomes. eLife. 2016;5:e11473. doi: 10.7554/eLife.11473. https://elifesciences.org/articles/11473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tibayrenc M., Ayala F.J. Reproductive clonality of pathogens: A perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa. Proc. Natl. Acad. Sci. USA. 2012;109(48):E3305–E3313. doi: 10.1073/pnas.1212452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tibayrenc M., Ayala F.J. How clonal are Trypanosoma and Leishmania? Trends Parasitol. 2013;29(6):264–269. doi: 10.1016/j.pt.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Ramirez J.D., Llewellyn M.S. Reproductive clonality in protozoan pathogens--truth or artefact? Mol. Ecol. 2014;23(17):4195–4202. doi: 10.1111/mec.12872. [DOI] [PubMed] [Google Scholar]
- 13.Ramirez J.D., Llewellyn M.S. Response to Tibayrenc and Ayala: Reproductive clonality in protozoan pathogens--truth or artefact? Mol. Ecol. 2015;24(23):5782–5784. doi: 10.1111/mec.13442. [DOI] [PubMed] [Google Scholar]
- 14.Messenger L.A., Miles M.A. Evidence and importance of genetic exchange among field populations of Trypanosoma cruzi. Acta Trop. 2015;151:150–155. doi: 10.1016/j.actatropica.2015.05.007. http://europepmc.org/ articles/PMC4644990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tibayrenc M., Ayala F.J. Reproductive clonality in protozoan pathogens--truth or artifact? A comment on Ramirez and Llewellyn. Mol. Ecol. 2015;24(23):5778–5781. doi: 10.1111/mec.13443. [DOI] [PubMed] [Google Scholar]
- 16.Gibson W. Liaisons dangereuses: Sexual recombination among pathogenic trypanosomes. Res. Microbiol. 2015;166(6):459–566. doi: 10.1016/j.resmic.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Akopyants N.S., Kimblin N., Secundino N., Patrick R., Peters N., Lawyer P., Dobson D.E., Beverley S.M., Sacks D.L. Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science. 2009;324(5924):265–268. doi: 10.1126/science.1169464. http://science.sciencemag.org/content/324/ 5924/265.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomasini N., Lauthier J.J., Ayala F.J., Tibayrenc M., Diosque P. How often do they have sex? A comparative analysis of the population structure of seven eukaryotic microbial pathogens. PLoS One. 2014;9(7):e103131. doi: 10.1371/journal.pone.0103131. journals.plos.org /plosone/article?id=10.1371/journal.pone.0103131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rougeron V., De Meeus T., Kako Ouraga S., Hide M., Banuls A.L. 2010 doi: 10.1371/journal.ppat.1001004. http://journals.plos.org/plospathogens/article?id=10.1371/ [DOI] [PMC free article] [PubMed]
- 20.Truc P., Buscher P., Cuny G., Gonzatti M.I., Jannin J., Joshi P., Juyal P., Lun Z.R., Mattioli R., Pays E., Simarro P.P., Teixeira M.M., Touratier L., Vincendeau P., Desquesnes M. Atypical human infections by animal trypanosomes. PLoS Negl. Trop. Dis. 2013;7(9):e2256. doi: 10.1371/journal.pntd.0002256. journals.plos.org/plosntds /article?id=10.1371/journal.pntd.0002256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaunt M.W., Yeo M., Frame I.A., Stothard J.R., Carrasco H.J., Taylor M.C., Mena S.S., Veazey P., Miles G.A., Acosta N., de Arias A.R., Miles M.A. Mechanism of genetic exchange in American trypanosomes. Nature. 2003;421(6926):936–939. doi: 10.1038/nature01438. https://www.nature.com/articles/nature01438 [DOI] [PubMed] [Google Scholar]
- 22.Roellig D.M., Savage M.Y., Fujita A.W., Barnabe C., Tibayrenc M., Steurer F.J., Yabsley M.J. Genetic variation and exchange in Trypanosoma cruzi isolates from the United States. PLoS One. 2013;8(2):e56198. doi: 10.1371/journal.pone.0056198. journals.plos.org /plosone/article?id=10.1371/journal.pone.0056198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomasini N., Diosque P. Evolution of Trypanosoma cruzi: Clarifying hybridisations, mitochondrial introgressions and phylogenetic relationships between major lineages. Mem. Inst. Oswaldo Cruz. 2015;110(3):403–413. doi: 10.1590/0074-02760140401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomasini N., Lauthier J.J., Monje Rumi M.M., Ragone P.G., Alberti D’Amato A.M., Brandan C.P., Basombrio M.A., Diosque P. Preponderant clonal evolution of Trypanosoma cruzi I from Argentinean Chaco revealed by Multilocus Sequence Typing (MLST). Infect. Genet. Evol. 2014;27:348–354. doi: 10.1016/j.meegid.2014.08.003. www.sciencedirect.com/science/article/pii/S1567134814002901 [DOI] [PubMed] [Google Scholar]
- 25.Messenger L.A., Llewellyn M.S., Bhattacharyya T., Franzen O., Lewis M.D., Ramirez J.D., Carrasco H.J., Andersson B., Miles M.A. Multiple mitochondrial introgression events and heteroplasmy in Trypanosoma cruzi revealed by maxicircle MLST and next generation sequencing. PLoS Negl. Trop. Dis. 2012;6(4):e1584. doi: 10.1371/journal.pntd.0001584. journals.plos.org/plosntds/article?id= 10.1371/journal.pntd.0001584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez J.D., Guhl F., Messenger L.A., Lewis M.D., Montilla M., Cucunuba Z., Miles M.A., Llewellyn M.S. Contemporary cryptic sexuality in Trypanosoma cruzi. Mol. Ecol. 2012;21(17):4216–4226. doi: 10.1111/j.1365-294X.2012.05699.x. [DOI] [PubMed] [Google Scholar]
- 27.Ocana-Mayorga S., Llewellyn M.S., Costales J.A., Miles M.A., Grijalva M.J. Sex, subdivision, and domestic dispersal of Trypanosoma cruzi lineage I in southern Ecuador. PLoS Negl. Trop. Dis. 2010;4(12):e915. doi: 10.1371/journal.pntd.0000915. journals.plos.org/plosntds /article?id=10.1371/journal.pntd.0000915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brisse S., Henriksson J., Barnabe C., Douzery E.J., Berkvens D., Serrano M., De Carvalho M.R., Buck G.A., Dujardin J.C., Tibayrenc M. Evidence for genetic exchange and hybridization in Trypanosoma cruzi based on nucleotide sequences and molecular karyotype. Infect. Genet. Evol. 2003;2(3):173–183. doi: 10.1016/s1567-1348(02)00097-7. [DOI] [PubMed] [Google Scholar]
- 29.Lewis M.D., Llewellyn M.S., Gaunt M.W., Yeo M., Carrasco H.J., Miles M.A. Flow cytometric analysis and microsatellite genotyping reveal extensive DNA content variation in Trypanosoma cruzi populations and expose contrasts between natural and experimental hybrids. Int. J. Parasitol. 2009;39(12):1305–1317. doi: 10.1016/j.ijpara.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnabe C., Breniere S.F. Scarce events of mitochondrial introgression in Trypanosoma cruzi: New case with a Bolivian strain. Infect. Genet. Evol. 2012;12(8):1879–1883. doi: 10.1016/j.meegid.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 31.Lewis M.D., Llewellyn M.S., Yeo M., Acosta N., Gaunt M.W., Miles M.A. Recent, independent and anthropogenic origins of Trypanosoma cruzi hybrids. PLoS Negl. Trop. Dis. 2011;5(10):e1363. doi: 10.1371/journal.pntd.0001363. journals.plos.org/plosntds/article?id= 10.1371/journal.pntd.0001363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukes J., Guilbride D.L., Votypka J., Zikova A., Benne R., Englund P.T. Kinetoplast DNA network: Evolution of an improbable structure. Eukaryot. Cell. 2002;1(4):495–502. doi: 10.1128/EC.1.4.495-502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruvalcaba-Trejo L.I., Sturm N.R. 2011 doi: 10.1186/1471-2164-12-58. central.com/articles/10.1186/1471-2164-12-58 [DOI] [PMC free article] [PubMed]
- 34.Aphasizheva I., Aphasizhev R. U-insertion/deletion mRNA-editing holoenzyme: Definition in sight. Trends Parasitol. 2016;32(2):144–156. doi: 10.1016/j.pt.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Degrave W., Fragoso S.P., Britto C., van Heuverswyn H., Kidane G.Z., Cardoso M.A., Mueller R.U., Simpson L., Morel C.M. Peculiar sequence organization of kinetoplast DNA minicircles from Trypanosoma cruzi. Mol. Biochem. Parasitol. 1988;27(1):63–70. doi: 10.1016/0166-6851(88)90025-4. [DOI] [PubMed] [Google Scholar]
- 36.Jensen R.E., Englund P.T. Network news: The replication of kinetoplast DNA. Annu. Rev. Microbiol. 2012;66:473–491. doi: 10.1146/annurev-micro-092611-150057. https://jhu.pure.elsevier.com/en/publications/network-news-the-replication-of-kinetoplast-dna-5 [DOI] [PubMed] [Google Scholar]
- 37.Klingbeil M.M., Drew M.E., Liu Y., Morris J.C., Motyka S.A., Saxowsky T.T., Wang Z., Englund P.T. Unlocking the secrets of trypanosome kinetoplast DNA network replication. Protist. 2001;152(4):255–262. doi: 10.1078/1434-4610-00066. [DOI] [PubMed] [Google Scholar]
- 38.Gibson W., Crow M., Kearns J. Kinetoplast DNA minicircles are inherited from both parents in genetic crosses of Trypanosoma brucei. Parasitol. Res. 1997;83(5):483–488. doi: 10.1007/s004360050284. [DOI] [PubMed] [Google Scholar]
- 39.Machado C.A., Ayala F.J. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc. Natl. Acad. Sci. USA. 2001;98(13):7396–7401. doi: 10.1073/pnas.121187198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Freitas J.M., Augusto-Pinto L., Pimenta J.R., Bastos-Rodrigues L., Goncalves V.F., Teixeira S.M., Chiari E., Junqueira A.C., Fernandes O., Macedo A.M., Machado C.R., Pena S.D. 2006 doi: 10.1371/journal.ppat.0020024. journals.plos.org/plospathogens/article?id=10.1371/journal.ppat [DOI] [PMC free article] [PubMed]
- 41.Zingales B., Andrade S.G., Briones M.R., Campbell D.A., Chiari E., Fernandes O., Guhl F., Lages-Silva E., Macedo A.M., Machado C.R., Miles M.A., Romanha A.J., Sturm N.R., Tibayrenc M., Schijman A.G. A new consensus for Trypanosoma cruzi intraspecific nomenclature: Second revision meeting recommends TcI to TcVI. Mem. Inst. Oswaldo Cruz. 2009;104(7):1051–1054. doi: 10.1590/s0074-02762009000700021. [DOI] [PubMed] [Google Scholar]
- 42.Zingales B., Miles M.A., Campbell D.A., Tibayrenc M., Macedo A.M., Teixeira M.M., Schijman A.G., Llewellyn M.S., Lages-Silva E., Machado C.R., Andrade S.G., Sturm N.R. The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect. Genet. Evol. 2012;12(2):240–253. doi: 10.1016/j.meegid.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 43.Flores-Lopez C.A., Machado C.A. 2011. plos.org/plosntds/article?id=10.1371/journal.pntd.0001272
- 44.El-Sayed N.M. 2005. [Google Scholar]
- 45.Brisse S., Barnabe C., Tibayrenc M. Identification of six Trypanosoma cruzi phylogenetic lineages by random amplified polymorphic DNA and multilocus enzyme electrophoresis. Int. J. Parasitol. 2000;30(1):35–44. doi: 10.1016/s0020-7519(99)00168-x. [DOI] [PubMed] [Google Scholar]
- 46.Westenberger S.J., Barnabe C., Campbell D.A., Sturm N.R. Two hybridization events define the population structure of Trypanosoma cruzi. Genetics. 2005;171(2):527–543. doi: 10.1534/genetics.104.038745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sturm N.R., Vargas N.S., Westenberger S.J., Zingales B., Campbell D.A. Evidence for multiple hybrid groups in Trypanosoma cruzi. Int. J. Parasitol. 2003;33(3):269–279. doi: 10.1016/s0020-7519(02)00264-3. [DOI] [PubMed] [Google Scholar]
- 48.Sturm N.R., Campbell D.A. Alternative lifestyles: The population structure of Trypanosoma cruzi. Acta Trop. 2010;115(1-2):35–43. doi: 10.1016/j.actatropica.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 49.Martin D.P., Posada D., Crandall K.A., Williamson C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retroviruses. 2005;21(1):98–102. doi: 10.1089/aid.2005.21.98. [DOI] [PubMed] [Google Scholar]
- 50.Franco J., Ferreira R.C., Ienne S., Zingales B. 2015 doi: 10.1016/j.meegid.2015.01.030. http://www.sciencedirect.com/science/article/pii/ [DOI] [PubMed]
- 51.Ferreira R.C., Briones M.R. Phylogenetic evidence based on Trypanosoma cruzi nuclear gene sequences and information entropy suggest that inter-strain intragenic recombination is a basic mechanism underlying the allele diversity of hybrid strains. Infect. Genet. Evol. 2012;12(5):1064–1071. doi: 10.1016/j.meegid.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 52.Tomazi L., Kawashita S.Y., Pereira P.M., Zingales B., Briones M.R. Haplotype distribution of five nuclear genes based on network genealogies and Bayesian inference indicates that Trypanosoma cruzi hybrid strains are polyphyletic. Genet. Mol. Res. 2009;8(2):458–476. doi: 10.4238/vol8-2gmr591. [DOI] [PubMed] [Google Scholar]
- 53.Ienne S., Pedroso A., Carmona E.F., Briones M.R., Zingales B. Network genealogy of 195-bp satellite DNA supports the superimposed hybridization hypothesis of Trypanosoma cruzi evolutionary pattern. Infect. Genet. Evol. 2010;10(5):601–606. doi: 10.1016/j.meegid.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 54.Bryant D., Moulton V. Neighbor-net: An agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 2004;21(2):255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- 55.Tomasini N., Diosque P. Phylogenomics of Trypanosoma cruzi: Few evidence of TcI/TcII mosaicism in TcIII challenges the hypothesis of an ancient TcI/TcII hybridization. Infect. Genet. Evol. 2017;50:25–27. doi: 10.1016/j.meegid.2017.02.010. www.sciencedirect.com/science/ article/pii/S1567134817300527 [DOI] [PubMed] [Google Scholar]
- 56.Cribb P., Tapia E., Diosque P., Serra E. Spliced leader RNA gene promoter sequence heterogeneity in CL-Brener Trypanosoma cruzi reference strain. Infect. Genet. Evol. 2004;4(2):153–157. doi: 10.1016/j.meegid.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 57.Elias M.C., Vargas N., Tomazi L., Pedroso A., Zingales B., Schenkman S., Briones M.R. Comparative analysis of genomic sequences suggests that Trypanosoma cruzi CL Brener contains two sets of non-intercalated repeats of satellite DNA that correspond to T. cruzi I and T. cruzi II types. Mol. Biochem. Parasitol. 2005;140(2):221–227. doi: 10.1016/j.molbiopara.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 58.Diosque P., Tomasini N., Lauthier J.J., Messenger L.A., Monje Rumi M.M., Ragone P.G., Alberti-D’Amato A.M., Perez Brandan C., Barnabe C., Tibayrenc M., Lewis M.D., Llewellyn M.S., Miles M.A., Yeo M. Optimized multilocus sequence typing (MLST) scheme for Trypanosoma cruzi. PLoS Negl. Trop. Dis. 2014;8(8):e3117. doi: 10.1371/journal.pntd.0003117. journals.plos.org /plosntds/article?id=10.1371/journal.pntd.0003117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Machado C.A., Jousselin E., Kjellberg F., Compton S.G., Herre E.A. Phylogenetic relationships, historical biogeography and character evolution of fig-pollinating wasps. Proc. Biol. Sci. 2001;268(1468):685–694. doi: 10.1098/rspb.2000.1418. http://www.stri.si.edu/sites/ publications/PDFs/STRI-W_Herre_Machado_etal_2001.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marcili A., Lima L., Cavazzana M., Junqueira A.C., Veludo H.H., Maia Da Silva F., Campaner M., Paiva F., Nunes V.L., Teixeira M.M. A new genotype of Trypanosoma cruzi associated with bats evidenced by phylogenetic analyses using SSU rDNA, cytochrome b and Histone H2B genes and genotyping based on ITS1 rDNA. Parasitology. 2009;136(6):641–655. doi: 10.1017/S0031182009005861. [DOI] [PubMed] [Google Scholar]
- 61.Lauthier J.J., Tomasini N., Barnabe C., Rumi M.M., D’Amato A.M., Ragone P.G., Yeo M., Lewis M.D., Llewellyn M.S., Basombrio M.A., Miles M.A., Tibayrenc M., Diosque P. Candidate targets for multilocus sequence typing of Trypanosoma cruzi: Validation using parasite stocks from the Chaco Region and a set of reference strains. Infect. Genet. Evol. 2012;12(2):350–358. doi: 10.1016/j.meegid.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 62.Torralba D., Baixauli F., Sanchez-Madrid F. Mitochondria know no boundaries: Mechanisms and functions of intercellular mitochondrial transfer. Front. Cell Dev. Biol. 2016;4:107. doi: 10.3389/fcell.2016.00107. https://www.frontiersin.org/articles/10.3389/fcell.2016.00107 /full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lynch M. Mutation accumulation in transfer RNAs: Molecular evidence for Muller’s ratchet in mitochondrial genomes. Mol. Biol. Evol. 1996;13(1):209–220. doi: 10.1093/oxfordjournals.molbev.a025557. [DOI] [PubMed] [Google Scholar]
- 64.Maciver S.K. Asexual Amoebae escape Muller’s ratchet through polyploidy. Trends Parasitol. 2016;32(11):855–862. doi: 10.1016/j.pt.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 65.Currat M., Ruedi M., Petit R.J., Excoffier L. The hidden side of invasions: Massive introgression by local genes. Evolution. 2008;62(8):1908–1920. doi: 10.1111/j.1558-5646.2008.00413.x. [DOI] [PubMed] [Google Scholar]
- 66.Neiva J., Pearson G.A., Valero M., Serrao E.A. Surfing the wave on a borrowed board: range expansion and spread of introgressed organellar genomes in the seaweed Fucus ceranoides L. Mol. Ecol. 2010;19(21):4812–4822. doi: 10.1111/j.1365-294X.2010.04853.x. [DOI] [PubMed] [Google Scholar]