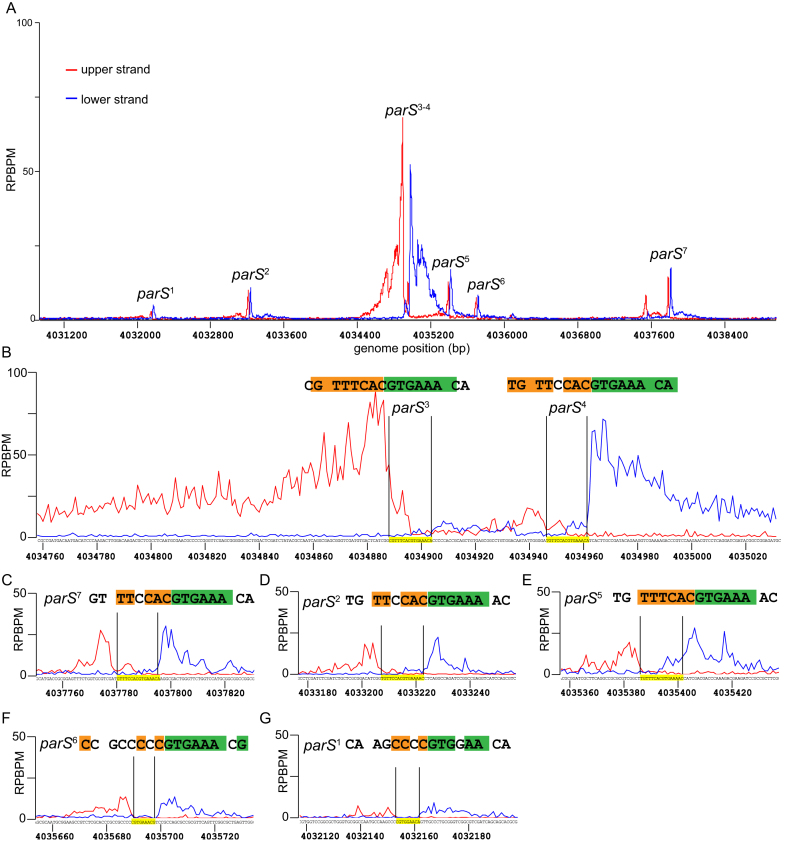
Figure 2.
Identification of parS sequences by in vitro DNA purification with deep sequencing (IDAP-seq). Sequencing reads were sorted to either the upper DNA strand (red) or to the lower strand (blue) of the Caulobacter reference gnome, as suggested in the original IDAP-seq publication (31). The sequence in between the summit of the upper strand profile and that of the lower strand profile defines the parS sequence required for binding to ParB in vitro (see also Supplementary Figure S3). (A) IDAP-seq profile of ParB-(His)6 in the genomic region between +4031 kb and +4039 kb. (B–G) IDAP-seq profile of ParB-(His)6 surrounding each individual parS site. Palindromic nucleotides within the identified parS site are shaded in orange and green.