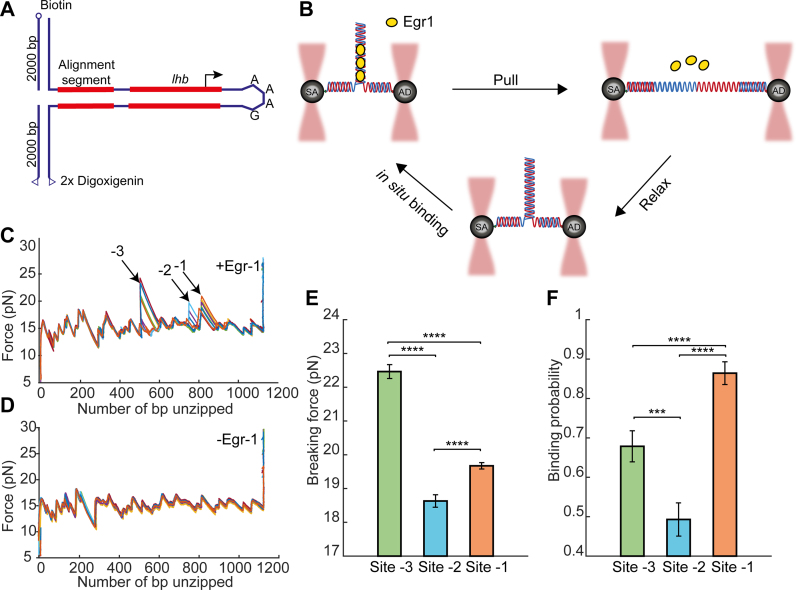
Figure 1.
Single-molecule probing of Egr-1 binding to the Lhb promoter. (A) The −517/+246 segment of mouse Lhb DNA is ligated to a fixed 350 bp alignment sequence and a short stem-loop that prevents breaking of the tether after unzipping. (B) The Lhb DNA is connected to two 2-kb dsDNA molecular handles, which are attached to polystyrene beads trapped in two separate optical traps. Egr-1 binding to the construct is initiated in situ (inside the experimental chamber). One of the traps is moved to stretch the tethered construct and disrupt protein–DNA interactions. After unzipping the whole construct, the DNA is relaxed and dsDNA forms again. The process is repeated multiple times. (C and D) Unzipping curves of Lhb in the presence (C) and absence (D) of Egr-1. Binding to each of the sites (-1, -2 and -3) is designated with arrows. (E) Breaking force for the three Egr-1 binding sites located on the Lhb promoter. Data shown as mean ± s.e.m., n = 140; ****P < 0.0001, two-sample Student’s t-test. (F) Binding probability, calculated as the number of binding events out of the total number of DNA unzipping cycles, for each of the three Egr-1 binding sites at [Egr-1] = 500 nM. Data shown as fraction ± s.e., n = 140; ****P < 0.0001, ***P < 0.001, Chi-square test.