Figure 7.
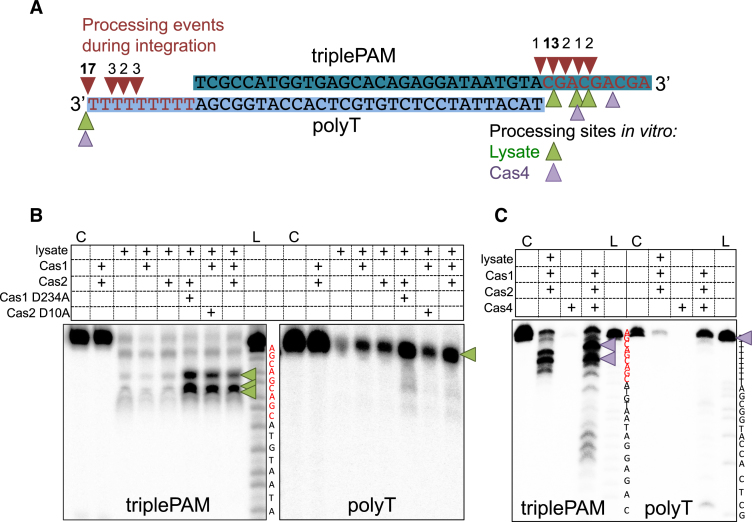
Prespacer processing is influenced by PAM sequences. (A) A prespacer with 3 PAM complementary sequences (5′-CGA-3′) in the 3′ single-stranded region of one strand (triplePAM) and a poly-T at the other 3′ end (polyT) (triplePAM_prespacer (Table 1) was integrated into pCRISPR A in a SPIN assay containing S. solfataricus cell lysate. The products of integration of either strand at site 1 were cloned and sequenced to identify processing events at the 3′ end of the prespacers. All processing events occurred in the 3′ overhangs and are indicated by red arrows, the number above indicates how many integration events were processed at each site. Green arrows indicate processing sites following incubation with Cas1–2AB and cell lysate (see B), and purple arrows indicate processing in the presence of Cas1–2AB and Cas4 (Sso1391) (see C). (B) The prespacer used in A was also incubated with Cas1–2AB, 5 mM MnCl2 and S. solfataricus lysate at 60°C and products run on a 15% denaturing urea–TBE polyacrylamide gel. The left hand panel shows the result of labeling the triplePAM strand and the right the result of labeling the polyT strand. The first lane is a control with only the labeled substrate loaded, followed by the products of incubation with Cas1–2AB or Cas proteins in combination with S. solfataricus (Sso) lysate. Inactive mutant D234A Cas1 or D10A Cas2 were also included in incubations as indicated. An A+G ladder (L) was also loaded to map the products of triplePAM processing. Green arrows indicate the major processing products. (C) The same prespacer was assayed with Cas1–2AB and cell lysate or Cas4 (Sso1391) to compare processing. The first lane is a control without protein (C) and an A+G ladder (L) was also loaded for each labelled species. Purple arrows indicate the major processing products.