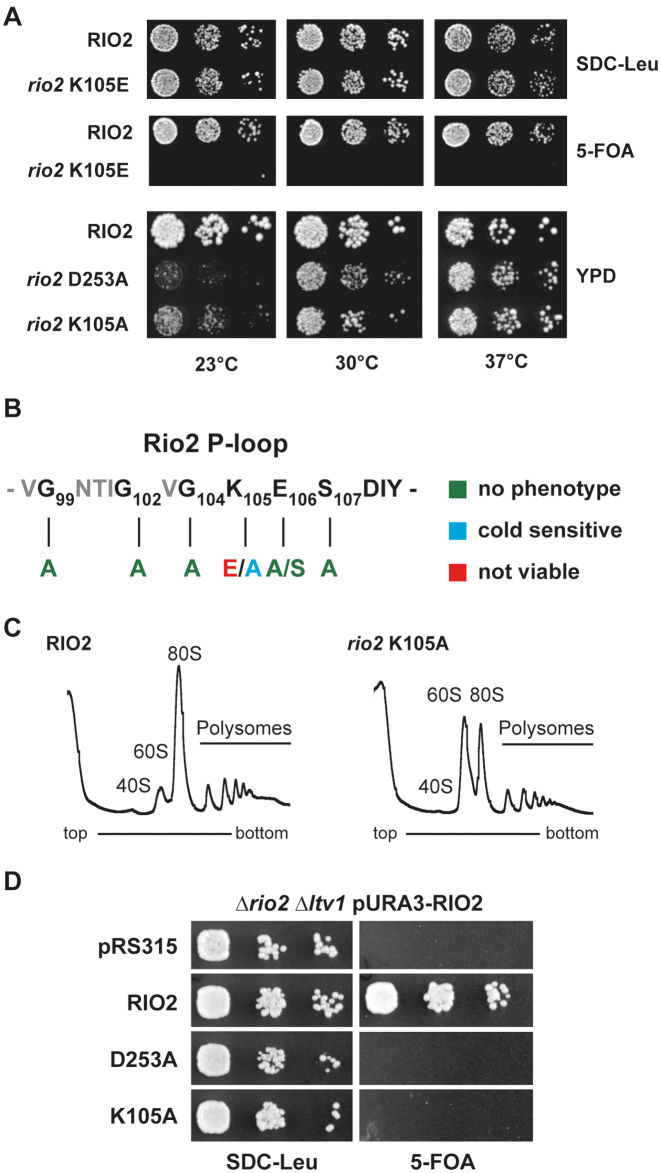
Figure 5.
P-loop lysine is necessary for optimal growth at low temperature and SSU biogenesis. (A) In vivo analysis of P-loop lysine mutants. Serial dilutions of yeast strains expressing wild-type RIO2 or the indicated mutations were spotted on the indicated plates and incubated at the indicated temperatures. RIO2 shuffle strain transformed with LEU2-carrying plasmids harboring wild-type Sc_RIO2 or Sc_rio2 K105E, were spotted on SDC-Leu and SDC plus 5-FOA plates (to shuffle out wild-type pURA3-RIO2). Yeast strains solely expressing wild-type RIO2 or the indicated mutations Sc_rio2 D253A and K105A were spotted on YPD. (B) Summary of phenotypes observed after P-loop mutagenesis. The phenotypes observed after individually mutating the following Sc_Rio2 P-loop residues G99A, G102A, G104A, K105A, K105E, E106A, E106S, S107A and S107E are summarized. (C) Polysome profile analysis of the Sc_rio2 K105A mutant. Whole cell lysates from cells incubated for 4 h at 23°C were separated on a 5–40% sucrose gradient. The A260nm profiles of the derived sucrose gradient fractions are depicted. (D) Genetic interaction of Sc_rio2 K105A allele with the non-essential ribosome biogenesis factor Ltv1. Serial dilutions of yeast strains expressing wild-type RIO2 or the indicated mutations were spotted on the indicated plates and incubated at the indicated temperatures. RIO2 shuffle strain deleted for LTV1 and transformed with LEU2-carrying plasmids harboring wild-type and mutated Rio2, were spotted on SDC-Leu and SDC plus 5-FOA plates.