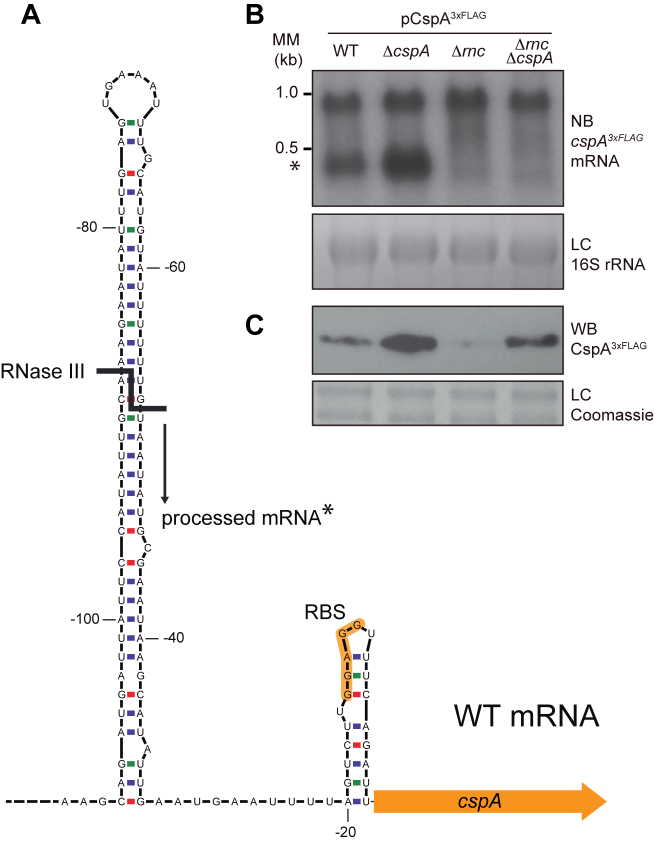
Figure 5.
Processing of cspA mRNA by RNase III. (A) Schematic representation of the cspA 5′UTR stem loop. Adapted from Lioliou and co-workers (30). The RNase III processing site is represented by a bold line and the resulting processed mRNA is indicated with an asterisk. (B) cspA3xFLAG mRNA levels produced from the pCspA3xFLAG plasmid in the WT, ΔcspA, Δrnc and ΔrncΔcspA strains. MM, Millenium Marker, band lengths are indicated. (C) CspA3xFLAG protein levels produced from these same strains. Total RNA and protein extraction were performed after growth until mid-exponential phase at 37°C and 200 rpm. RNA samples were run into 1.25% agarose and transferred to Nitran membranes. Northern blots were then developed using a 32P-labeled anti-FLAG oligo probe and autoradiography. Ethidium bromide staining of 16S RNA is shown as loading control (LC). Protein samples were run into 12% polyacrylamide gels and transferred to nitrocellulose membranes. Western blots were then developed using peroxidase conjugated anti-FLAG antibodies and bioluminescence kit. Coomassie stained gel portions are shown as loading controls (LC).