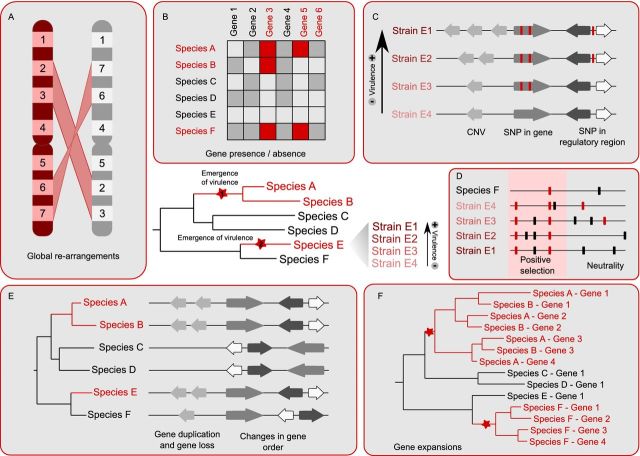
Figure 1.
Schematic overview of comparative genomics methods to find virulence factors. In the center of the image a phylogenetic tree represents the evolutionary relationships between an idealized group of pathogenic and non-pathogenic species, where the ones drawn in red are pathogenic. Red stars indicate two points in the tree where virulence emerged. For species E, a list of strains is indicated on the right showing the presence of sequenced strains with different degrees of virulence. The boxes surrounding the tree indicate different kinds of analyses that can be performed to detect virulence traits. (A) Search for genomic re-arrangements. (B) Presence and absence of genes, shown as a table where the white squares represent missing genes and the gray and red square represent present genes. In addition, red squares indicate putative virulence factors. (C) Detection of differences between strains. Each arrow represents a gene. Red horizontal lines represent SNPs. (D) Detection of positive selection. Red horizontal lines represent non-synonymous SNPs and black horizontal lines represent synonymous SNPs. (E) Events that can be detected with the use of phylogenetic trees. Arrows represent genes. Gene duplication and loss is represented along with changes in gene order. (F) Gene expansions. Phylogenetic tree representing two independent expansion events in the pathogenic species. Expansion points are marked with a star.