Figure 2.
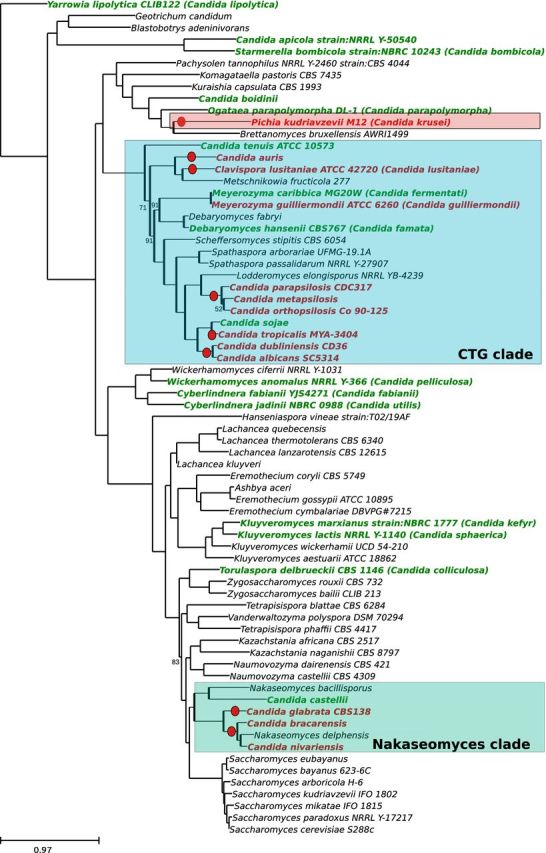
Phylogenetic relationships among Candida. A total of 516 genes detected as single copy genes in Saccharomyces species (Marcet-Houben and Gabaldón 2015) were used to perform a homology search against a proteome database formed by the 71 Saccharomycotina species included in the tree. A phylogenetic tree was reconstructed for each group of homologs using the phylome reconstruction pipeline (Huerta-Cepas et al.2014). Phylogenetic trees were examined with ETE (Huerta-Cepas, Dopazo and Gabaldón 2010), species-specific duplications were deleted and one sequence belonging to the duplicated clade was randomly chosen to represent the clade. Trees that, after filtering, contained one-to-one orthologs in at least three-fourth of the species were retained and their alignments were concatenated. The final alignment contained 190 110 amino acid positions. The phylogenetic tree was reconstructed using RAxML with the PROTGAMMALG model (Stamatakis, Ludwig and Meier 2005). Bootstraps were reconstructed using RAxML rapid bootstrap approach, supports below 100 are marked on the tree. For Candida species, red-colored leaves indicate pathogenic species while green-colored leaves indicate non-pathogenic species.
