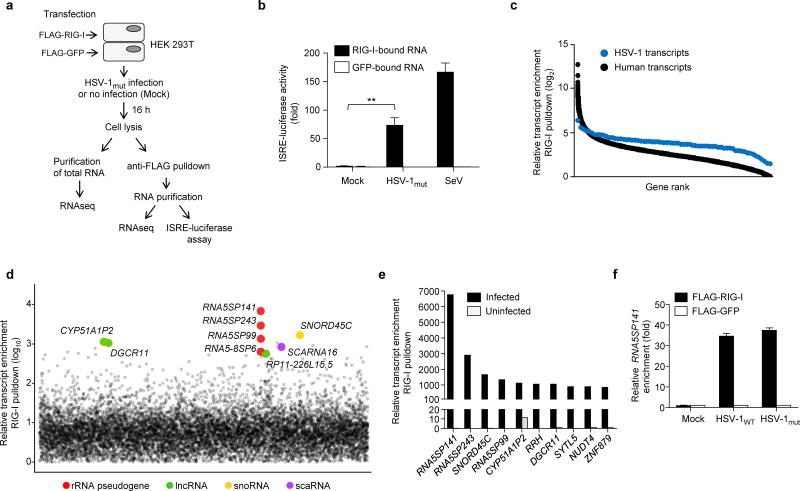
Figure 1.
Endogenous non-coding RNAs co-immunoprecipitate with RIG-I during HSV-1 infection. (a) Schematic representation of the experimental setup for isolation and identification of RNAs from FLAG-RIG-I- or FLAG-GFP-precipitates. RNAseq, next-generation RNA sequencing. (b) ISRE-luciferase reporter activity in HEK 293T cells transfected for 18 h with 7 μl RNA from FLAG-RIG-I- or FLAG-GFP-precipitates from uninfected (Mock) or HSV-1mut-infected cells, isolated as described in (a). RNA from FLAG-RIG-I- or FLAG-GFP-precipitates from HEK 293T cells infected with SeV (50 HAU/ml) served as control. Luciferase activity is presented as fold induction relative to the values for FLAG-GFP-precipitates from uninfected cells, set to 1. (c) Relative enrichment of HSV-1–derived or human-derived transcripts in FLAG-RIG-I precipitates from HSV-1mut-infected cells, determined by RNAseq. Relative enrichment (log2) of transcripts was calculated by comparing the abundance of transcripts in FLAG-RIG-I precipitates to FLAG-GFP precipitates. Data are from two independent experiments. (d) Relative enrichment (log10) of human transcripts in FLAG-RIG-I precipitates from HSV-1mut-infected cells. The 9 most highly-enriched non-coding transcripts are shown. rRNA pseudogene (red); long non-coding RNA (lncRNA, green); small nucleolar RNA (snoRNA, yellow); small Cajal body-specific RNAs (scaRNA, purple). (e) Relative enrichment of the 10 most highly-enriched human transcripts (both non-coding and coding) in FLAG-RIG-I precipitates from HSV-1mut-infected cells, as compared to uninfected cells. (f) Quantitative RT-PCR (qRT-PCR) analysis of RNA5SP141 transcripts from RNA isolated as described in (a) from HEK 293T cells transfected with FLAG-RIG-I or FLAG-GFP and infected with HSV-1WT or HSV-1mut (both MOI 1), or left uninfected (Mock). Data are representative of two independent experiments (b,f; mean and s.d. of n = 3 technical replicates) or are from two independent experiments (c-e). **P < 0.005 (unpaired t-test).