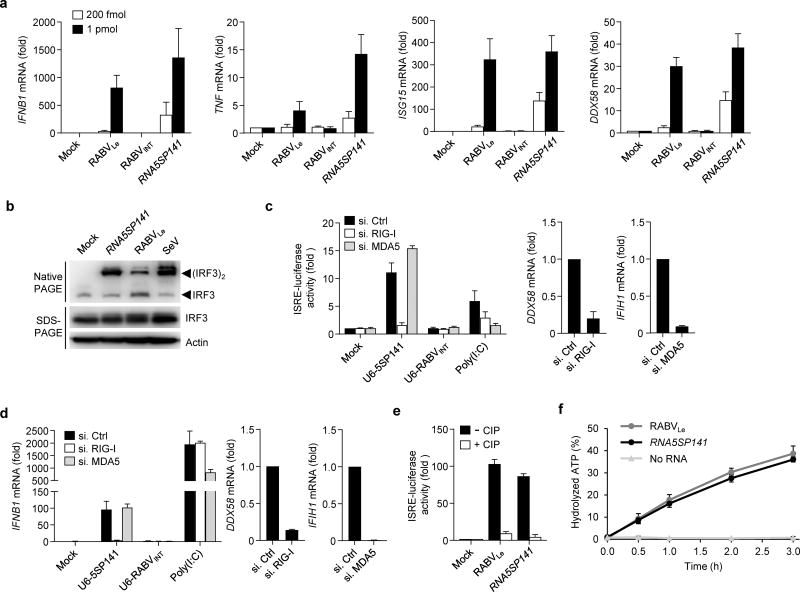
Figure 2.
RNA5SP141 activates RIG-I. (a) qRT-PCR analysis of the indicated cytokine and ISG transcripts (vertical axes) in HEK 293T cells transfected with 200 fmol or 1 pmol in vitro-transcribed RNA5SP141, or in vitro–transcribed RNA corresponding to the rabies virus leader sequence (RABVLe; positive control) or an internal rabies virus sequence (RABVINT; negative control). (b) Native PAGE and SDS-PAGE of lysates from HEK 293T cells transfected with 125 pmol of the indicated in vitro transcripts or infected with 50 HAU/ml SeV for 16 h, or left untreated (Mock). Endogenous IRF3 and actin were detected by immunoblot (IB) with anti-IRF3 and anti-actin, respectively. (c) Left: ISRE-luciferase reporter activity in HEK 293T cells transfected for 30 h with non-targeting control siRNA (si.Ctrl) or siRNAs targeting RIG-I or MDA5 (si.RIG-I or si.MDA5), and subsequently mock-transfected or transfected with 500 ng of a DNA construct encoding U6 promoter-expressed RNA5SP141 or RABVINT (U6-5SP141 or U6-RABVINT) for 18 h. Treatment with 1 µg/ml HMW-poly(I:C) served as a control. Middle, right: Knockdown efficiency of endogenous RIG-I (DDX58) and MDA5 (IFIH1) was confirmed by qRT-PCR. (d) Left: qRT-PCR analysis of IFNB1 mRNA in NHLF cells that were transfected with the indicated siRNAs and U6 promoter-expressed DNA constructs as in (c). Treatment with 0.05 µg/mL HMW-poly(I:C) served as a control. Middle, right: Knockdown efficiency of endogenous RIG-I (DDX58) and MDA5 (IFIH1) was confirmed by qRT-PCR. (e) ISRE-luciferase reporter activity in HEK 293T cells transfected for 18 h with 500 fmol in vitro–transcribed RNA5SP141 or RABVLe, which had been pre-treated with calf alkaline phosphate (CIP), or left untreated. (f) Quantification of hydrolyzed [γ-32P]ATP by RIG-I incubated for the indicated times with 250 nM of in vitro-transcribed RNA5SP141. Incubation of RIG-I with in vitro–transcribed RABVLe, or no RNA, served as positive and negative controls, respectively. Free phosphate was separated from unhydrolyzed ATP by thin layer chromatography, and the percentage of hydrolyzed ATP in each sample was calculated. Data are representative of two (b–f) or three (a) independent experiments (mean and s.d. of n = 2 biological replicates in a, n = 3 biological replicates in b–e, n = 3 technical replicates in f).