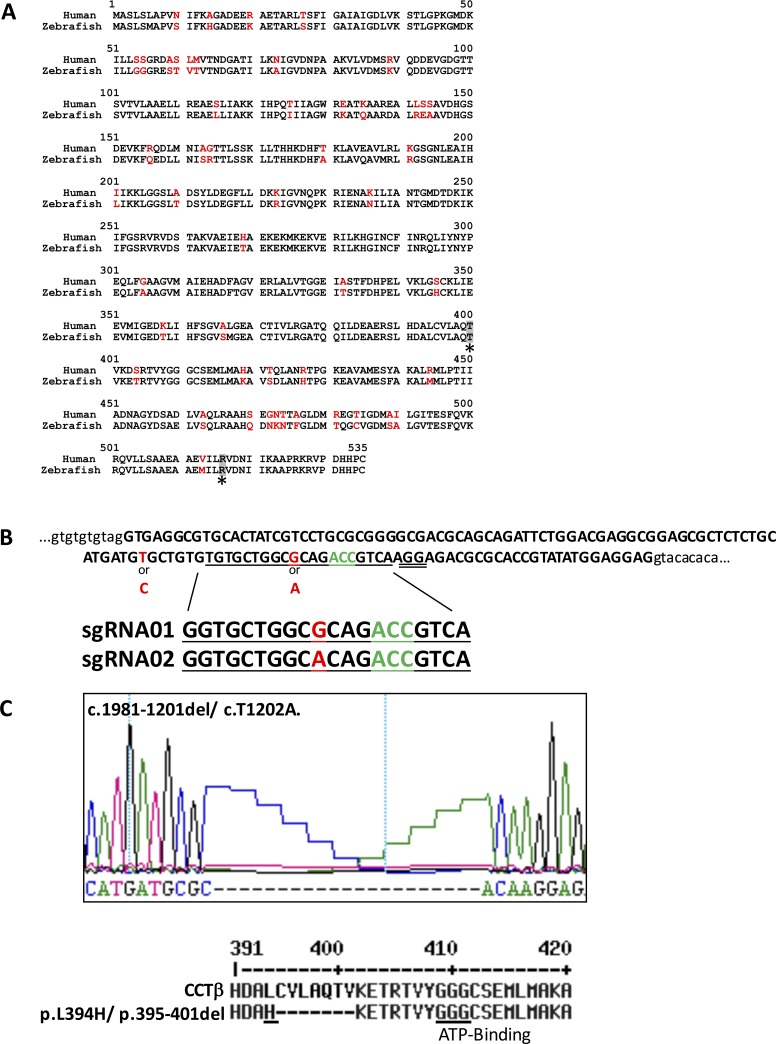
Figure 1.
Strategy and summary of the genome editing for cct2 by CRISPR-Cas9. (A) Comparison of human and zebrafish CCTβ amino acid sequences. The differing amino acid residues are shown in red. The position of LCA-associated mutations T400 and R516 are shaded and marked by asterisks. (B) The reference sequence of the targeted area of the zebrafish cct2 gene in the TL strain. Exon 14 of cct2 is depicted in capital letters. The intronic sequences surrounding this exon are depicted in lower case letters. The guide RNA sequences (underlined in the reference sequence) are shown below the reference sequence. T, the first 5′ nucleotide in the guide RNA sequences, was substituted by G for in vitro transcription using T7 polymerase. The polymorphic positions are shown in red. The PAM sequence was double underlined. The codon corresponding to threonine 400 is shown in green. (C) Changes in the nucleotide and protein sequences of the cct2-L394H-7del mutant. Upper panel shows the nucleotide sequence. The lower panel shows translated wild-type and mutated proteins. The mutated CCTβ protein has the L394H substitution (underlined) and deletion of seven amino acids. The highly conserved ATP-binding motif is underlined.