Abstract
The direct and efficient conversion of alcohols into amines is a pivotal transformation in chemistry. Here, we present an artificial, oxidation–reduction, biocatalytic network that employs five enzymes (alcohol dehydrogenase, NADP-oxidase, catalase, amine dehydrogenase and formate dehydrogenase) in two concurrent and orthogonal cycles. The NADP-dependent oxidative cycle converts a diverse range of aromatic and aliphatic alcohol substrates to the carbonyl compound intermediates, whereas the NAD-dependent reductive aminating cycle generates the related amine products with >99% enantiomeric excess (R) and up to >99% conversion. The elevated conversions stem from the favorable thermodynamic equilibrium (K’eq = 1.88 × 1042 and 1.48 × 1041 for the amination of primary and secondary alcohols, respectively). This biocatalytic network possesses elevated atom efficiency, since the reaction buffer (ammonium formate) is both the aminating agent and the source of reducing equivalents. Additionally, only dioxygen is needed, whereas water and carbonate are the by-products. For the oxidative step, we have employed three variants of the NADP-dependent alcohol dehydrogenase from Thermoanaerobacter ethanolicus and we have elucidated the origin of the stereoselective properties of these variants with the aid of in silico computational models.
Introduction
The direct conversion of alcohol moieties into the corresponding amines is a pivotal transformation in chemistry as a great number of chemical products and intermediates contain amine functionalities (e.g. active pharmaceutical ingredients, agrochemicals, fine chemicals, polymers, dyes, plasticizers, emulsifiers, detergents, etc.).1 Chemical and enzymatic methods are currently widely applied in industry for the conversion of prochiral ketones into optically active amines.2,3 However, a future transition to a bio-based economy requires the development of efficient strategies for the direct conversion of alcohols into amines. A classical method for the one-pot synthesis of amines from alcohols4 relies on three sequential steps: (i) the conversion of alcohol into azide via Mitsunobu reaction,5 (ii) the transformation of azide to iminophosphorane via Staudinger reaction,6 followed by (iii) hydrolysis to the amine product in diluted acid. However, this method and related ones possess extremely low atom efficiency because of the requirement of various and complex reagents in stoichiometric amounts. Organometal-catalysis using iridium, ruthenium, iron or copper complexes enables the one-pot amination of alcohols using a single catalyst in a process that is known as hydrogen-borrowing amination.7,8 This method encompasses a first oxidation step of the alcohol into a carbonyl compound followed by the in situ generation of an imine intermediate and its reduction to the amine. In this process, the hydride liberated in the first step is quantitatively consumed in the second step. Nevertheless, the chemocatalytic hydrogen-borrowing amination using ammonia as amine donor is plagued by low chemoselectivity because complex mixtures of primary, secondary and tertiary amines are obtained.9–12 Modifications of the chemocatalytic hydrogen-borrowing amination with improved chemoselectivity require other amine donors such as aniline and its derivatives,13,14 or amino-pyridines,15 or an enantiopure auxiliary.16 Nonetheless, the moderate or total absence of stereoselectivity, the elevated catalyst and cocatalyst loading, the requirement of an excess of the alcohol starting material at a low concentration, the stringent reaction conditions and the observed substrate inactivation of the catalyst during operation constitute current challenges that thwart the widespread application of these methods.17
In this context, the exquisite properties of compatibility, chemo- and stereoselectivity of enzymes allowed our group and others to develop recently the asymmetric biocatalytic hydrogen-borrowing amination of alcohols (Scheme 1A).18–20 In the first step an alcohol dehydrogenase (ADH)21,22 oxidizes the alcohol substrate to the carbonyl compound intermediate with concomitant transfer of the hydride to the oxidized form of the nicotinamide coenzyme (NAD+). In the second step, an amine dehydrogenase (AmDH)3,23–28 performs the reductive amination of the carbonyl moiety employing the NADH, which was generated in the first cycle, and ammonia. Under the optimized reaction conditions,29 secondary alcohols were converted up to 95% conversion within 48 h. Another formal enzymatic hydrogen-borrowing amination utilizes a combination of an ADH, a ω-transaminase and an alanine dehydrogenase.30–34 However, the latter method has lower efficiency due to the requirement of 5 equivalents of alanine as additional sacrificial amine donor and results in modest conversion and mediocre selectivity for the amination of secondary alcohols.
Scheme 1.
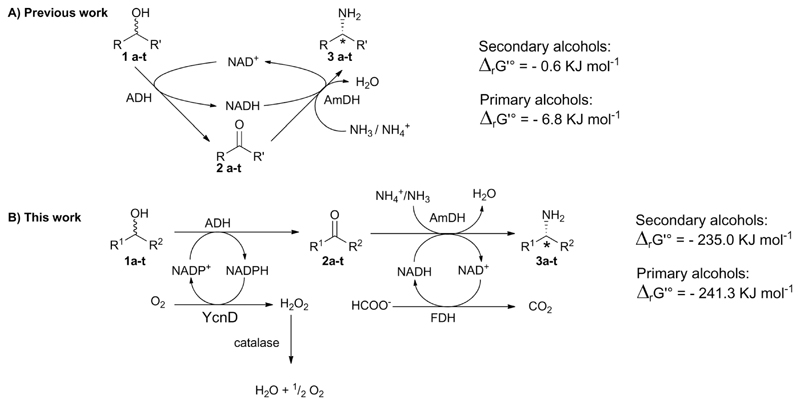
Biocatalytic strategies for the one-pot conversion of alcohols to enantiopure amines employing alcohol dehydrogenases (ADHs) and amine dehydrogenases (AmDHs): (A) biocatalytic hydrogen-borrowing amination (i.e. redox-interconnected steps); (B) orthogonal biocatalytic network (i.e. concurrent redox separated steps, this work). ΔrG’° is the calculated free Gibbs energy for the reaction in standard conditions (T 298.15 K, P 1 bar) and in aqueous buffer at pH 8.5, ionic strength 1 M.
In this work, we present a new and high efficient one-pot biocatalytic strategy for the asymmetric amination of secondary and primary alcohols. The new method relies on the different specificity of some oxidoreductases for accepting either the phosphorylated form (NADP) or the non-phosphorylated form (NAD) of the nicotinamide coenzyme (Scheme 1B). In the first oxidative cycle, an ADH oxidizes the alcohol starting material to the carbonyl compound intermediate (e.g. ketone or aldehyde) by the transfer of a hydride from the substrate to NAPD+. The highly specific NADP-oxidase (YcnD) from Bacillus subtilis35 regenerates in situ NADP+, consuming dioxygen and liberating H2O2. A third enzyme, a catalase, effects the disproportion of H2O2 to water and dioxygen; the latter re-enters in the first cycle. In the second reductive cycle, an amine dehydrogenase (AmDH) performs the reductive amination of the carbonyl compound intermediate to afford the final amine product. A formate dehydrogenase from Candida boidinii (Cb-FDH)36 recycles the required NADH at the expense of formate. Notably, the reaction buffer (e.g. ammonium formate) is both the source of ammonium and electrons for the reductive amination step, whereas the electron acceptor in the first step is the innocuous molecular oxygen from air. As the two cycles are running concurrently in one-pot without compartmentalization, the biocatalytic network constitutes an elegant example of an oxidation–reduction orthogonal chemical process.37 Only one other similar type of biocatalytic orthogonal two-step oxidation–reduction network (i.e. involving a combination of dehydrogenases with divergent cofactor dependence, NAD or NADP) has been created previously for the deracemisation of racemic alcohols.38,39 Another outstanding application of the same concept for the deracemization of alcohols required, instead, the compartmentalization of the enzymes in order to separate physically the oxidative and the reductive steps.40
Therefore, the herein reported oxidation–reduction biocatalytic network for the amination of alcohols is conceptually different from other already known systems. In fact, other types of oxidation–reduction cascades for the deracemization of alcohol moieties were created through the combination of an alcohol oxidase (e.g. Cu2+ or flavin dependent) for the oxidative step, with a dehydrogenase for the reductive step.41,42 A laccase/TEMPO system can also substitute the alcohol oxidase in the oxidative step.43 Finally, the combination of an alcohol oxidase, a transaminase along with an alanine dehydrogenase for recycling of the amine donor, allowed for the one-pot two-step amination of alcohols.44,45 The variation of the latter cascade using a laccase/TEMPO system with a transaminase has been recently published.46 The last example is the orthogonal two-step amination of alcohols involving ω-transaminases, which was limited by a maximum of 32% conversion.31
Results and discussion
Thermodynamic calculations
As alcohol and amine are in the same oxidation state, no additional reagents are required in the biocatalytic hydrogen-borrowing amination (Scheme 1A). Therefore, the hydrogen-borrowing amination possesses the highest atom efficiency. On the other hand, the biocatalytic hydrogen-borrowing amination is a reversible reaction; hence, the final conversion is dictated by the thermodynamics of the reaction. In fact, the variation of the standard Gibbs free energy for the biocatalytic hydrogen-borrowing amination of secondary alcohols was estimated in this work as ΔrG′° −0.6 kJ mol−1 (for details, see ESI Table S1†).47 This value was calculated for an aqueous system, at pH 8.5 and ionic strength 1 M. These parameters are analogous to the experimental conditions applied in our biocatalytic hydrogen borrowing amination.18 Using the same approach, the estimated ΔrG′° for the amination of primary alcohols is −6.8 kJ mol−1 (for details, see ESI Table S2†). These data are in agreement with our previous experimental results that showed a more favorable thermodynamic equilibrium for the hydrogen-borrowing amination of primary alcohols (>99% conversion) compared to secondary alcohols (max 95% conversion) using an excess of ammonium/ammonia as driving force for the reaction.
Thus, we thought that an oxidation–reduction orthogonal chemical process (Scheme 1B) might lead, in general, to quantitative conversions because of a more favorable thermodynamic equilibrium. Furthermore, the use of two separated redox steps may also lead to higher reaction rates. Our calculations showed that the estimated overall ΔrG′° for the amination of secondary alcohols following the orthogonal pathway depicted in Scheme 1b is −235.0 kJ mol−1 (i.e. K′eq = 1.48 × 1041, for details see ESI Table S5†). Also in this case, the amination of primary alcohols is more favored as the estimated overall ΔrG′° is −241.3 kJ mol−1 (i.e. K′eq = 1.88 × 1042, for details see ESI Table S8†). Interestingly, the thermodynamic driving force mainly stems from the first oxidative step (for details, see ESI paragraph 5†).
Testing suitable alcohol dehydrogenases and rational understanding of their stereoselective properties
Aiming at creating an atom-efficient and orthogonal biocatalytic network for the amination of secondary alcohols with improved thermodynamic equilibrium, we initially searched for suitable high selective NADP-dependent enzymes for the oxidative cycle.
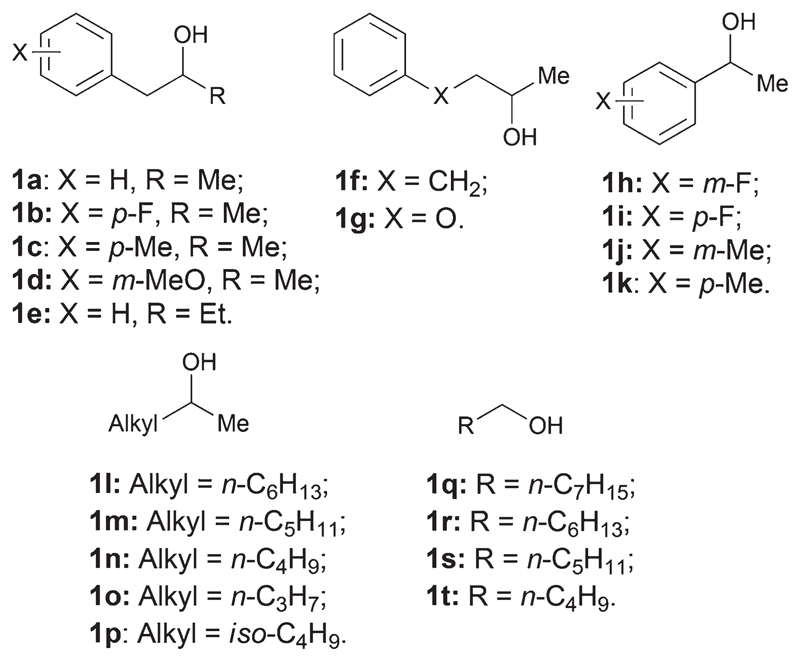
The alcohol substrates selected for our study are reported in Fig. 1.
Fig. 1.
Structures of the secondary (1a–p) and primary (1q–t) alcohol substrates explored in this study. For the detailed structural representation of the substrates, see ESI Fig. S1–3.†
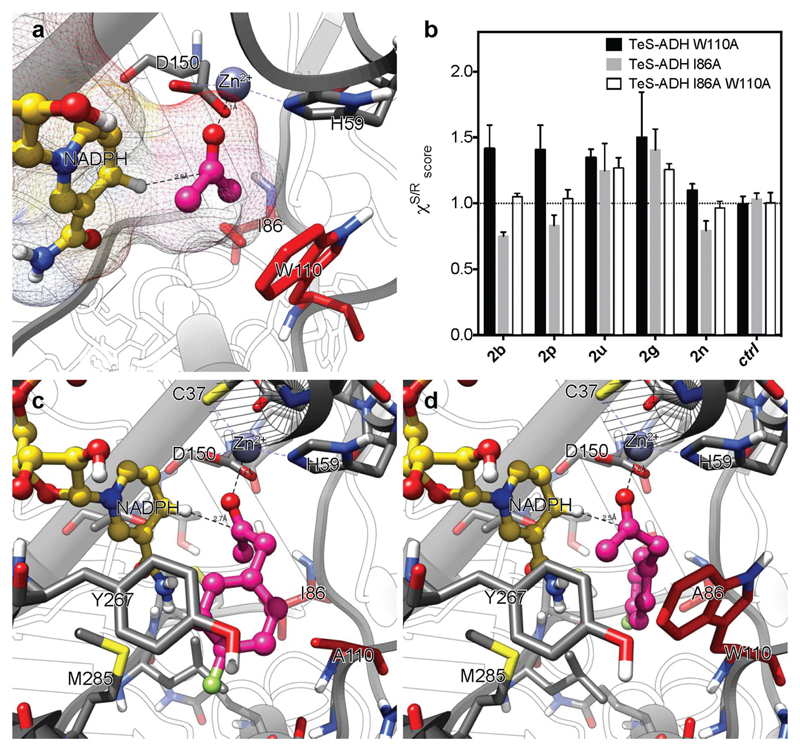
Besides the specificity for NADP as coenzyme, the amination of S or R configured secondary alcohols as well as their racemic mixtures requires the availability of stereocomplementary ADHs or a non-enantioselective ADH. In this regard, a Prelog ADH from Thermoanaerobacter ethanolicus (TeS-ADH, also known as ADH-T) was previously characterized as NADP-dependent ADH; however, its substrate scope was limited to the reduction of short and medium-length chain aliphatic ketones.48–52 The mutation of tryptophan 110 into an alanine (TeS-ADH W110A variant) has shown to increase the substrate scope of the ADH to include a number of aromatic ketones.53–55 In another study of enzyme engineering on the same scaffold, the mutation of isoleucine 86 into an alanine created a second variant (TeS-ADH I86A) that showed reversed stereoselectivity (e.g. anti-Prelog) for the reduction of a set of seven aromatic and heteroaromatic ketones.56 Furthermore, other variants were created via: (i) saturation mutagenesis of the amino acid residue W110;57,58 (ii) combination of the mutations I86A, W110A and/or inclusion of a further mutation such as C295A.58–60 A number of these variants showed relaxed stereoselectivity for the reduction of prochiral ketones and, therefore, they were applied for the deracemization of alcohols or dynamic kinetic resolution.58,61 Nevertheless, comprehensive data on the activity and stereoselectivity of TeS-ADH W110A and TeS-ADH I86A towards our selection of alcohol substrates were not available. Thus, we investigated the reactivity and the stereoselectivity of TeS-ADH W110A and TeS-ADH I86A for the asymmetric reduction of structurally diverse ketones (enantiomeric excesses are reported in ESI, Tables S9 and S10†). We expressed the ADHs with a Strep-II-tag at the C-terminus that allows for an easier purification. As expected, TeS-ADH W110A afforded always the Prelog alcohol (S-configured alcohol, for the selected substrates) as the major enantiomer. Nonetheless, the enzyme was stereospecific (enantiomeric excess, ee ≥99%) only in four cases out of thirteen. In particular, the reduction of 2-hexanone (2n) afforded a nearly racemic mixture of alcohols (5% ee) whereas the reduction of 2-heptanone (1m) gave the (S)-enantiomer in 39% ee. These findings are in agreement with previous publications that showed the imperfect stereoselectivity of TeS-ADH W110A.53,54,62 Conversely, our biocatalytic orthogonal aminating network utilizes the ADH to accomplish the oxidation of an alcohol substrate to a ketone intermediate. Thus, TeS-ADH W110A turned out to be a useful catalyst because perfect stereoselectivity is not required for our purpose: the important property is instead the elevated selectivity of TeS-ADH W110A for the coenzyme (i.e. NADP compared to NAD).
Then, we investigated whether TeS-ADH I86A is indeed a general anti-Prelog variant because the literature reports the reductions of only few ketones as substrates.56 Surprisingly, we discovered that TeS-ADH I86A is a non-stereospecific enzyme when tested with our selection of substrates (see ESI, Table S10†). TeS-ADH I86A always afforded mixtures of enantiomers that were enriched either of the Prelog (S-configured) or anti-Prelog (R-configured) alcohol. In particular, reverse stereoselectivity compared to TeS-ADH W110A (i.e. main formation of the R-enantiomer) was observed only for the reduction of para-fluoro phenylacetone (2b), 2-hexanone (2n) and 4-methyl butan-2-one (2p). In all the other cases investigated, the S-enantiomer (i.e. Prelog) was still the main product with an enantiomeric excess ranging from 27% to >99%. Hence, TeS-ADH I86A cannot be applied as a general anti-Prelog ADH in our orthogonal biocatalytic network. Therefore, we turned our attention to the anti-Prelog ADH from Lactobacillus brevis (Lb-ADH).63 A wealth of data regarding the substrate scope and the stereoselectivity of Lb-ADH for the reduction of prochiral ketones in analytical and preparative scale is available from literature.64–67 In all the cases reported, it was shown that Lb-ADH affords the anti-Prelog alcohol as the major enantiomer with excellent enantiomeric excess. Thus, it was not required to investigate the reduction of ketones with Lb-ADH any further.
The final extension of our biocatalytic network to racemic alcohols as starting material requires a non-enantioselective ADH in the first oxidative step. It has been reported that simultaneous mutations of WT TeS-ADH at the positions I86, W110 and C295 with alanine residues generates variants with relaxed enantioselectivity.59 However, C295 might be involved in the binding of the catalytic Zn2+ ion of the TeS-ADH. Therefore, mutation of C295 might decrease the stability of TeS-ADH as previously observed by other authors.59 This finding is corroborated further by our computational models (see below), which show a distance of 4 Å between the thiol group of C295 and the catalytic Zn2+. As robust enzymes are generally preferable for a concurrent multi-step artificial biocatalytic network, we focused our interest to TeS-ADH I86A W110A double variant.58,68 We expressed the enzyme in our lab as Strep-II-tagged recombinant enzyme and we evaluated its stereoselectivity as for TeS-ADH W110A and TeS-ADH I86A. TeS-ADH I86A W110A was completely non-stereoselective (ee 0%) for the reduction of 2-hexanone (2n). Additionally, an extremely low level of stereoselectivity (i.e. from 10% to 24%) was observed for the reduction of aryl-aliphatic ketones 2a, 2b, 2d and all the other aliphatic ketones 2m–p (see ESI, Table S15†). The origin of the stereoselective properties of the three variants (TeS-ADH W110A, TeS-ADH I86A and TeS-ADH I86A W110A) and the related intricacies of the amino acid residues I86 and W110 were elucidated via computational studies. Local docking studies on the mutant TeS-ADH W110G with 2-tetralone and derivatives were published concomitantly to our study.68 First, we created a series of models for TeS-ADH W110A, TeS-ADH I86A and TeS-ADH I86A W110A based on the structure of the thermophilic alcohol dehydrogenase from Thermoanaerobacter brockii as the template (PDB code 1YKF).69 Structural and computational studies for Tb-ADH are also available.70 Notably, the wild-type ADH from Thermoanaerobacter ethanolicus (WT TeS-ADH) and the ADH from Thermoanaerobacter brockii (Tb-ADH) possess both 352 amino acid residues and they differ only in three of them (i.e. 99% structural identity, ESI paragraph 13†). Furthermore, the three different amino acid residues are located further from the active site (for more information see Experimental part). Herein, the 3D model structures of TeS-ADHs (WT, W110A, I86A and I86A W110A) were created containing acetone as model substrate in the active site. Afterwards, acetone was utilized as a pro-chiral control in all the model structures of the TeS-ADH variants because all the substrates were in silico created from acetone. We performed this control in order to ensure that the enzymatic system under study does not show any chirality biases (i.e. pseudo-prochirality preferences for a totally symmetric substrate). In practice, we also submitted the enzymes models studied herein to the MD protocol using acetone as ligand. Observing the simulation, such chirality biases did not occur. Hence, starting from the reactive pose of acetone in the active site of all TeS-ADH variants, models with bound substrates 2b (para-fluorophenylacetone), 2p (4-methylpentan-2-one), 2u (acetophenone), 2g (phenoxy-acetone), and 2n (2-hexanone) were obtained (see Experimental part for detailed procedure). This selection of the substrates was made to study all the possible cases: (i) low ee values for TeS-ADH I86A W110A, but high ee values for TeS-ADH W110A and TeS-ADH I86A W110A (2b and 2p); (ii) high ee values for all the TeS-ADH variants (2u and 2g); (iii) low or moderate ee values for all the TeS-ADH variants (2n). We point out that the model of WT TeS-ADH was used as a starting point for the generation of the models of the TeS-ADH variants, but it was not considered for simulations any further.
Following the in silico protocol (see Experimental part), we were able to score the enantioselective preferences of these TeS-ADH variants as well as to understand the structural consequences of inducing mutations at the amino acid positions 86 and 110. One can observe in Fig. 1a that acetone was located in its reactive conformation in the active site of the model of WT TeS-ADH. In Fig. 2a, it can also be observed that the active site of WT TeS-ADH is rather constricted in space, hence limiting the substrate acceptance to rather non-voluminous substrates such as acetone. This finding is in agreement with the early reported substrate scope of the WT enzyme that was limited to medium-length chain linear aliphatic ketones.49 As previously reported, I86 and W110 are located in opposite positions in the active site of the enzyme.55,56 Mutation of one of these positions to a rather small side chain amino acid such as alanine, would allow for the creation of space in the active site that is required to accommodate the substrate either in the pro-S or pro-R orientation. In the present work, these structural intricacies were experimentally determined and computationally confirmed by inducing such mutations. In relation to the stereoselective properties of the TeS-ADH variants, the level of agreement between the experimental data and the computational models was evaluated using a scoring function ( see the Experimental part for more details). A score value higher than 1 indicates a high preference for the pro-S binding conformation of the given substrate, a score value lower than 1 indicates higher preference for the pro-R binding conformation of the given substrate, while a score equal or close to 1 indicates no stereoselective preferences of the variant for the given substrate. The average score values for each variant and each selected substrate is reported in Fig. 2b. It is noteworthy that these theoretical results showed a good agreement with the experimental data, hence validating the model structures presented in this work.
Fig. 2.
Structural intricacies of enantioselective preferences of TeS-ADH. Panel a shows the active site of WT TeS-ADH with acetone in its reactive conformation. Panel b shows the scoring values of all the TeS-ADH variants for substrates 2b, 2p, 2u, 2g, 2n and acetone, the latter used as a control substrate (ctrl). A score value higher than 1 means high preference for the pro-S binding conformation of the given substrate, a score value lower than 1 indicates higher preference for the pro-R binding conformation of the given substrate, while a score equal or close to 1 indicates no stereoselective preferences of the variant for the given substrate. Panel c and d show the active site of TeS-ADH W110A and TeS-ADH I86A, respectively, containing the substrate 2b in its pro-S conformation (panel c) and its pro-R conformation (panel d).
One can observe that the creation of more space into the “pro-S enzyme binding pocket” (W110A mutation in TeS-ADH W110A) allows a more voluminous substrates like 2b to be accommodated in the active site in a reactive conformation (see Fig. 2c). On the other hand, the generation of space in the opposite side (I86A mutation in TeS-ADH I86A) allows the same substrate to bind assuming its pro-R conformation (see Fig. 2d). Inducing both mutations at the same time (I86A and W110A mutations in TeS-ADH I86A W110A) generates space at both sides of the hydride transfer region of the enzyme, thus enabling the observed non-stereoselective preference of this variant towards most of the substrates from this study (ESI, Table S15†). It is interesting to note that TeS-ADH I86A W110A is still high pro-S selective for substrates 2u and 2g. This feature can be in part explained by the high tendency of these substrates to form a π–π interaction between their aromatic group and the residue Y267, which is located on the pro-S side of the hydride transfer center. Residue M285 seems to play a role as well.
Optimization of the oxidative step
We investigated the first oxidative cycle at different pH values. Hence, we combined the NADP-dependent W110A variant of the ADH from Thermoanaerobacter ethanolicus (TeS-ADH W110A) with a high coenzyme selective NADPH-oxidase from Bacillus subtilis (YcnD)35 to carry out the oxidation of (S)-phenyl 2-propanol ((S)-1a) to phenylacetone (2a). The following reaction conditions were applied: (S)-1a (20 mM), TeS-ADH W110A (13 μM), His6-tagged YcnD (5 μM), catalase (0.2 μM) and NADP+ (0.5 mM) in Tris-HCl buffer (50 mM) at varied pH (7–8.5). The most elevated conversion was 91% at pH 7.5 after 24 h reaction time, whereas performing the reaction at pH 8.5 led to 85% conversion (see ESI, Table S11,† entries 2 and 4, respectively). In a following set of experiments (see ESI, Table S12†) we tried to improve the conversion for the oxidation of (S)-1a in Tris-HCl buffer (50 mM) at pH 8.5 as it is the optimal pH for the reductive amination with AmDHs (i.e. second cycle in Scheme 1).18,24 In particular, at this stage, we had to apply a double concentration of TeS-ADH W110A (26 μM) and His6-YcnD (10 μM) and extend the reaction time up to 2 days in order to obtain 98% conversion. However, the use of HCOONH4 (the required buffer for the subsequent reductive amination) instead of Tris-HCl, under the same reaction conditions, reduced again the conversion (85% after 2 days, data not shown in ESI†).
In our previous study on the biocatalytic hydrogen-borrowing amination, we have shown that some ADHs (as purified enzymes in ammonium buffer) suffer of mediocre stability when combined in one-pot reactions with other enzymes that bear a poly-histidine tag.18 We proposed that the imidazole groups of the poly-histidine chain can coordinate to the divalent cations (e.g. Ca2+, Mg2+, Zn2+) that are present in the structure of most microbial ADHs. These cations are essential either for retaining the correct oligomeric state of the ADHs (i.e. stability) or for the catalytic activity. These properties apply to the ADHs used in the present study.55,63 In fact, when we used C-terminal His6-tagged YcnD for the dual-enzyme oxidation of (S)-1a, the formation of a white enzyme precipitate during the course of the biocatalytic oxidation was observed, hence corroborating our hypothesis. Consequently, we prepared an alternative recombinant YcnD that was devoid of a His-tag. Indeed, the substitution of C-terminal His6-tagged YcnD with the YcnD devoid of His-tag was beneficial for the oxidation as >99% conversion was obtained in HCOONH4 (1 M, pH 8.5) in 12 h under the optimized reaction conditions: (S)-1a (20 mM), TeS-ADH W110A (26 μM), YcnD (10 μM), catalase (0.2 μM) and NADP+ (0.5 mM), (data not shown in ESI†). This result confirms that one must consider and select very carefully enzyme terminal tags when planning multi-enzyme processes in vitro.18 For this reason, in this work, we expressed and purified the TeS-ADH variants with a Strep-II-tag linked at their C-terminus.
Orthogonal biocatalytic network for the amination of (S)-configured alcohols with inversion of configuration
The amine dehydrogenases (AmDHs) used in this study were obtained by protein engineering of wild-type amino acid dehydrogenases (AADHs) that are dependent on NAD as coenzyme. As none of the mutations was located in the binding site of the coenzyme, the AmDH variants retain the same specificity for NADH as the parent wild-type enzymes.25,28,71 This property is important in order to assure the orthogonality between the oxidative and the reductive cycle of the biocatalytic network.72 The AmDHs used in this study were: (i) Bb-PhAmDH that was originated from the phenylalanine dehydrogenase from Bacillus badius;71 (ii) Rs-PhAmDH that was originated from the phenylalanine dehydrogenase from Rhodococcus sp.;25 (iii) Ch1-AmDH that is a chimeric enzyme28 obtained via domain shuffling of Bb-AmDH and another variant originated from the leucine dehydrogenase from Geobacillus stereothermopilus.27 In a very recent extensive study we have determined the optimal conditions for the reductive amination in HCOONH4 buffer (1 M, pH 8.5) of carbonyl compounds (20–50 mM) catalyzed by AmDHs in combination with the formate dehydrogenase from Candida boidinii (Cb-FDH).24 The high specificity of Cb-FDH for the reduction of NAD+ to NADH is also very important for the present study.36
In a preliminary set of experiments (see ESI, Table S13†), we proved unequivocally that the presence of any terminal His-tagged enzyme in the biocatalytic network is detrimental. The amination of (S)-1b (20 mM) was attempted with Strep-II-tagged TeS-ADH W110A (26–52 μM), His-tagged YcnD (5–10 μM), His-tagged Ch1-AmDH (126 μM), Cb-FDH (20 μM) and catalase (0.2 μM) in HCOONH4 buffer (1 M, pH 8.5) at 30 °C. The concentration of both coenzymes NAD+ and NADP+ was 0.5 mM. The maximum conversion to the amine (R)-3b was limited to 64% after 48 h (ESI, Table S13,† entry 1); the remaining component of the reaction mixture was exclusively the alcohol (S)-1b (i.e. the concentration of ketone 2b was ca. 1%). Thus, the moderate conversion stems from a catalytic inefficiency of the ADH employed in the oxidation cycle. Other data (ESI, Table S13,† entries 1–4) revealed that a decrease in the conversion of (S)-1b into (R)-3b correlated with an increase of the concentration of His-tagged YcnD, independently from the concentration of Strep-II-tagged TeS-ADH W110A. Finally, the use of Ch1-AmDH devoid of His-tag in combination with His-tagged YcnD, slightly increased the conversion up to a maximum of 78% (ESI, Table S13,† comparing entries 1 and 5).
Finally, we repeated the biocatalytic reaction with His-tagged YcnD (5 µM) and His-tagged Ch1-AmDH (126 µM) by adding the Strep-II-tagged TeS-ADH W110A in small aliquots every three hours (4.5 μL per aliquot, 5 aliquots, total concentration 65 μM). In this case, the conversion raised up to 98% in 48 h. These data all together indicated that the limitation of the orthogonal biocatalytic network was due to the poor stability of the Strep-II-tagged TeS-ADH W110A in the concomitant presence of any other His-tagged enzyme in solution. Further experiments demonstrated that the activity of the Strep-II-tagged TeS-ADH W110A drops to undetectable levels within 1 h of incubation in presence of any other His-tagged enzyme from this study (data not shown).
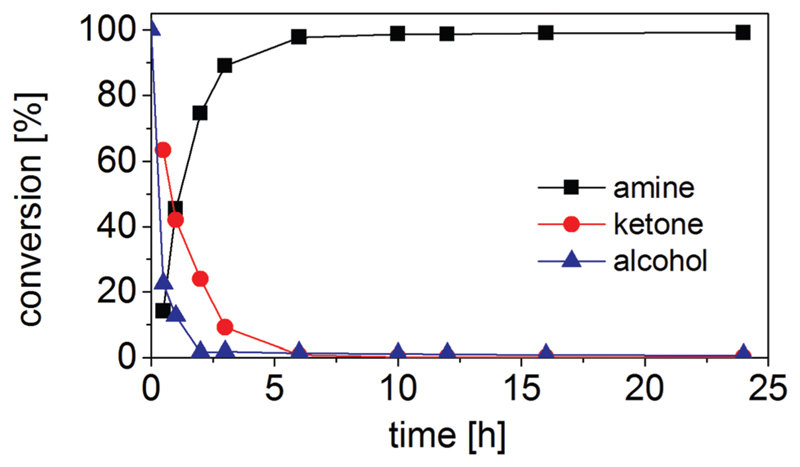
When we prepared all the five enzymes involved in the biocatalytic network as variants devoid of His-tags, the compatibility of the system was finally assured. Under the reaction conditions of (S)-1b (20 mM), Strep-II-tagged TeS-ADH W110A (45 μM), YcnD (10 μM), Ch1-AmDH (126 μM), Cb-FDH (20 μM), catalase (0.2 μM), NAD+ (0.5 mM) and NADP+ (0.5 mM) in HCOONH4 buffer (1 M, pH 8.5) at 30 °C, the concentration of the amine (R)-3b surpassed 99% after 12 h. Fig. 3 depicts the progress of the reaction over the time (ESI, Table S14†). The enantiomeric excess of the amine (R)-3b remained constant during the course of the reaction and it was more than 99%.
Fig. 3.
Time study for the amination of (S)-1b (20 mM) with inversion of configuration using the five-enzyme orthogonal biocatalytic network.
The broad applicability of the simultaneous two-step biocatalytic network was evaluated for the amination of a panel of (S)-configured secondary alcohols (Fig. 1). Since all the AmDHs from this study perform the amination of prochiral ketones with (R)-selectivity, the overall biocatalytic network proceeded with inversion of configuration.
(S)-Phenyl-propan-2-ol and derivatives ((S)-1a–d) were quantitatively converted into the related enantiopure amines (Table 1, entries 1–4) as predicted from our thermodynamic calculations. In addition, the other aryl-aliphatic alcohols ((S)-1e–g) were aminated with elevated conversion (95% or higher, Table 1, entries 5–7). In contrast, 1-phenylethanol and derivatives ((S)-1h–k) proved to be more challenging substrates as the conversion ranged from 17% to 22% (Table 1, entries 8–11). Depending on the substrate, we observed either a relevant amount of the alcohol starting material (2h, 2j) or an accumulation of the ketone intermediate (2i, 2k). In general, a favorable thermodynamic equilibrium is not the only requirement for achieving a quantitative amination within a reasonable time (e.g. ca. 24 h). In addition, both ADH and AmDH must possess relevant activity towards the alcohol substrate and ketone intermediate, respectively. Hence, the system is currently mainly limited by the low activity of the available AmDHs towards acetophenone and its derivatives as already reported in our previous publication.24 Additionally, the activity of the employed ADH is not ideal, yet, for the oxidation of (S)-2h and (S)-2j. Nonetheless, the final amine product was always obtained in enantiopure form. Finally, aliphatic secondary alcohols ((S)-1l–p) were in general well accepted affording up to >99% conversion (Table 1, entries 12–16). Notably, the orthogonal biocatalytic network proceeded with perfect stereoselectivity in all the cases investigated.
Table 1.
Orthogonal, oxidation–reduction, biocatalytic network for the amination of enantiopure secondary alcohols with inversion of configuration
| Enzymes |
Conversion [%] |
||||||
|---|---|---|---|---|---|---|---|
| Entry | Substrate | ADH | AmDH | Amine | Ketone | Alcohol | ee [%] |
| 1 | (S)-1a | TeS W110A | Ch1 | 99 | <1 | <1 | >99 (R) |
| 2 | (S)-1b | TeS W110A | Ch1 | 99 | <1 | <1 | >99 (R) |
| 3 | (S)-1c | TeS W110A | Rs | >99 | <1 | <1 | >99 (R) |
| 4 | (S)-1d | TeS W110A | Rs | >99 | <1 | <1 | >99 (R) |
| 5 | (S)-1e | TeS W110A | Rs | 95 | 1 | 4 | >99 (R) |
| 6 | (S)-1f | TeS W110A | Rs | >99 | <1 | <1 | >99 (R) |
| 7 | (S)-1g | TeS W110A | Bb | 95 | <1 | 5 | >99 (R) |
| 8 | (S)-1h | TeS W110A | Ch1 | 18 | 25 | 57 | >99 (R) |
| 9 | (S)-1i | TeS W110A | Ch1 | 22 | 78 | <1 | >99 (R) |
| 10 | (S)-1j | TeS W110A | Ch1 | 17 | 25 | 58 | >99 (R) |
| 11 | (S)-1k | TeS W110A | Ch1 | 22 | 73 | 5 | >99 (R) |
| 12 | (S)-1l | TeS W110A | Ch1 | 97 | 3 | <1 | >99 (R) |
| 13 | (S)-1m | TeS W110A | Ch1 | >99 | <1 | <1 | >99 (R) |
| 14 | (S)-1n | TeS W110A | Ch1 | >99 | <1 | <1 | >99 (R) |
| 15 | (S)-1o | TeS W110A | Ch1 | 72 | 21 | 8 | >99 (R) |
| 16 | (S)-1p | TeS W110A | Ch1 | 97 | 3 | <1 | >99 (R) |
Reaction conditions: S-Configured alcohol (20 mM), Strep-II-tagged TeS-ADH W110A (45 μM), YcnD (10 μM), Ch1-AmDH (126 μM) or Bb-PhAmDH (117 μM) or Rs-PhAmDH (100 μM), Cb-FDH (20 μM), catalase (0.2 μM), NAD+ (0.5 mM) and NADP+ (0.5 mM) in HCOONH4 buffer (1 M, pH 8.5) at 30 °C and orbital agitation (170 rpm).
Orthogonal biocatalytic network for the asymmetric amination of racemic alcohols
Finally, we applied the variant Strep-II-tagged TeS-ADH I86A W110A (52 μM) in the orthogonal biocatalytic network for the asymmetric amination of racemic alcohols. Starting from racemic 1a–b, 1l–n, 1p, enantiopure amines (>99% ee (R)) were obtained with high conversion (from 91% to >99%, Table 2, entries 1, 2, 13–15 and 17). Additionally, the amination of rac-1d and rac-1o afforded the optically pure amine product with 75% and 70% conversion, respectively (Table 2, entries 4 and 16). For the substrates rac-1c and rac-1e–k, it was required to combine two stereocomplementary NADP-dependent ADHs in one-pot: the Prelog TeS-ADH W110A and the anti-Prelog ADH from Lactobacillus brevis (Lb-ADH).63 The level of conversions were similar to the results obtained for the amination with inversion of configuration (Table 1). Aryl-aliphatic substrates were converted better (from 63% to >99%) than 1-phenylethanol derivatives (from 17% to 29%), (Table 2, entries 6–8 and entries 9–12, respectively). Finally, the amination of rac-1c–d afforded 53% and 91% conversion, respectively (Table 1, entries 3 and 5). The stereoselectivity was retained in all the cases (ee >99% (R)).
Table 2.
Orthogonal, oxidation–reduction, biocatalytic network for the asymmetric amination of racemic secondary alcohols
| Enzymes |
Conversion [%] |
||||||
|---|---|---|---|---|---|---|---|
| Entry | Substrate | ADH | AmDH | Amine | Ketone | Alcohol | ee [%] |
| 1 | Rac-1a | TeS I86A W110A | Ch1 | 97 | 3 | <1 | >99 (R) |
| 2 | Rac-1b | TeS I86A W110A | Ch1 | 91 | <1 | 9 | >99 (R) |
| 3 | Rac-1c | TeS W110A/Lb | Rs | 53 | <1 | 46 | >99 (R) |
| 4 | Rac-1d | TeS I86A W110A | Rs | 75 | <1 | 25 | >99 (R) |
| 5 | Rac-1d | TeS W110A/Lb | Rs | 91 | <1 | 9 | >99 (R) |
| 6 | Rac-1e | TeS W110A/Lb | Rs | 86 | 3 | 11 | >99 (R) |
| 7 | Rac-1f | TeS W110A/Lb | Rs | >99 | <1 | <1 | >99 (R) |
| 8 | Rac-1g | TeS W110A/Lb | Bb | 63 | <1 | 37 | >99 (R) |
| 9 | Rac-1h | TeS W110A/Lb | Ch1 | 29 | 30 | 41 | >99 (R) |
| 10 | Rac-1i | TeS W110A/Lb | Ch1 | 23 | 77 | <1 | >99 (R) |
| 11 | Rac-1j | TeS W110A/Lb | Ch1 | 21 | 23 | 56 | >99 (R) |
| 12 | Rac-1k | TeS W110A/Lb | Ch1 | 17 | 81 | 2 | >99 (R) |
| 13 | Rac-1l | TeS I86A W110A | Ch1 | 94 | 2 | 4 | >99 (R) |
| 14 | Rac-1m | TeS I86A W110A | Ch1 | >99 | <1 | <1 | >99 (R) |
| 15 | Rac-1n | TeS I86A W110A | Ch1 | >99 | <1 | <1 | >99 (R) |
| 16 | Rac-1o | TeS I86A W110A | Ch1 | 70 | 26 | 4 | >99 (R) |
| 17 | Rac-1p | TeS I86A W110A | Ch1 | 92 | 5 | 3 | >99 (R) |
Reaction conditions: Racemic alcohol (20 mM), Strep-II-tagged TeS-ADH I86A W110A (52 μM) or Strep-II-tagged TeS-ADH W110A (23 μM) plus Lb-ADH (23 μM), YcnD (10 μM), Ch1-AmDH (126 μM) or Bb-PhAmDH (117 μM) or Rs-PhAmDH (100 μM), Cb-FDH (20 μM), catalase (0.2 μM), NAD+ (0.5 mM) and NADP+ (0.5 mM) in HCOONH4 buffer (1 M, pH 8.5) at 30 °C and orbital agitation (170 rpm).
Furthermore, we performed the orthogonal biocatalytic network for the asymmetric amination of a racemic alcohol in preparative scale. Substrate 1m (50 mg) was aminated to afford the amine (R)-3m (>99% ee) with 97% conversion in 12 h. After a simple work-up, that is the extraction of the unreacted alcohol and ketone under acid conditions followed by the extraction of the amine under basic conditions, (R)-3m was isolated in pure form (45 mg, 90% yield, for details and product characterization see Experimental and ESI†). Notably, no purification (e.g. column chromatography) was required.
Orthogonal biocatalytic network for the amination of (R)-configured alcohols with retention of configuration
For the sake of completeness, we investigated the amination of enantiopure (R)-configured alcohols employing Lb-ADH. The results confirmed the general trend for the amination of aryl-aliphatic and aliphatic alcohols. However, the aminations of 1-phenylethanol derivatives afforded up to 43% conversion indicating that Lb-ADH is a more efficient ADH for the oxidation of these alcohols (Table 3).
Table 3.
Orthogonal, oxidation–reduction, biocatalytic network for the amination of enantiopure secondary alcohols with retention of configuration
| Entry | Substrate | Enzymes |
Conversion [%] |
ee [%] | |||
|---|---|---|---|---|---|---|---|
| ADH | AmDH | Amine | Ketone | Alcohol | |||
| 1 | (R)-1a | TeS I86A W110A | Rh | 96 | 1 | 3 | >99 (R) |
| 2 | (R)-1b | TeS I86A W110A | Rh | 96 | <1 | 4 | >99 (R) |
| 3 | (R)-1c | Lb | Rs | 30 | <1 | 70 | >99 (R) |
| 4 | (R)-1d | Lb | Rs | 6 | <1 | 94 | >99 (R) |
| 5 | (R)-1e | Lb | Rs | 97 | 1 | 2 | >99 (R) |
| 6 | (R)-1f | Lb | Rs | >99 | <1 | <1 | >99 (R) |
| 7 | (R)-1g | Lb | Bb | 82 | <1 | 18 | >99 (R) |
| 8 | (R)-1h | Lb | Ch1 | 43 | 10 | 47 | >99 (R) |
| 9 | (R)-1i | Lb | Ch1 | 28 | 72 | <1 | >99 (R) |
| 10 | (R)-1j | Lb | Ch1 | 36 | 40 | 24 | >99 (R) |
| 11 | (R)-1k | Lb | Ch1 | 20 | 73 | 7 | >99 (R) |
| 12 | (R)-1l | TeS I86A W110A | Ch1 | 79 | 2 | 19 | >99 (R) |
| 13 | (R)-1m | TeS I86A W110A | Ch1 | >99 | <1 | <1 | >99 (R) |
| 14 | (R)-1n | TeS I86A W110A | Ch1 | >99 | <1 | <1 | >99 (R) |
| 15 | (R)-1o | TeS I86A W110A | Ch1 | 80 | 18 | 2 | >99 (R) |
| 16 | (R)-1p | TeS I86A W110A | Ch1 | 89 | 2 | 9 | >99 (R) |
Reaction conditions: R-Configured alcohol (20 mM), Strep-II-tagged TeS-ADH I86A W110A (52 μM) or Lb-ADH (45 μM), YcnD (10 μM), Ch1-AmDH (126 μM) or Bb-PhAmDH (117 μM) or Rs-PhAmDH (100 μM), Cb-FDH (20 μM), catalase (0.2 μM), NAD+ (0.5 mM) and NADP+ (0.5 mM) in HCOONH4 buffer (1 M, pH 8.5) at 30 °C and orbital agitation (170 rpm).
Orthogonal biocatalytic network for the amination of primary alcohols
Terminal amines are also important compounds for the fine and bulk chemical industry.1 Hence, new sustainable routes for the conversion of primary alcohols to amines are of high interest. During tests for the activity of TeS-ADH I86A W110A, we found out that the variant is able to oxidize some primary alcohols. Consequently, we tested our orthogonal biocatalytic network for the amination of a small set of primary aliphatic alcohols applying the TeS-ADH I86A W110A in the first oxidative cycle. Medium-length chain alcohols as n-hexanol (1s) and n-heptanol (1r) were the most reactive substrates yielding the terminal amine with 88% and 75% conversion (Table 4, entries 2 and 3). Conversions dropped when we tested longer or shorter-length chain aliphatic alcohols (ca. 20%, Table 4, entries 1 and 4). In these latter cases, the aldehyde intermediate was not detected at the end of the reaction. This finding demonstrates that the oxidation of the alcohol substrate is currently the limiting step for the amination of 1q and 1t.
Table 4.
Orthogonal, oxidation–reduction, biocatalytic network for the amination of primary alcohols
| Enzymes |
Conversion [%] |
|||||
|---|---|---|---|---|---|---|
| Entry | Substrate | ADH | AmDH | Amine | Aldehyde | Alcohol |
| 1 | 1q | TeS I86A W110A | Ch1 | 20 | <1 | 80 |
| 2 | 1r | TeS I86A W110A | Ch1 | 75 | <1 | 25 |
| 3 | 1s | TeS I86A W110A | Ch1 | 88 | <1 | 13 |
| 4 | 1t | TeS I86A W110A | Ch1 | 19 | <1 | 81 |
Reaction conditions: Primary alcohol (20 mM), Strep-II tagged TeS-ADH I86A W110A (52 μM), YcnD (10 μM), Ch1-AmDH (126 μM), Cb-FDH (20 μM), catalase (0.2 μM), NAD+ (0.5 mM) and NADP+ (0.5 mM) in HCOONH4 buffer (1 M, pH 8.5) at 30 °C and orbital agitation (170 rpm).
Experimental
General optimized procedure for the orthogonal biocatalytic amination of enantiopure alcohols with inversion of configuration on analytical scale
The reactions were conducted in ammonium formate buffer (1 M, pH 8.5, final volume 0.5 mL) containing NADP+ and NAD+ (final concentration 0.5 mM each). Enzymes Strep-II-tagged TeS-ADH W110A (45 µM), YcnD (10 µM), AmDH (either Ch1-AmDH 126 µM or Bb-AmDH 117 µM or Rs-AmDH 100 µM), Cb-FDH (20 µM) and catalase (0.2 μM) and the substrate (50 mM) were added. Finally (S)-configured alcohol substrate (20 mM) was added. The reactions were run at 30 °C in an incubator for 24 h (170 rpm). Work-up was performed by the addition of KOH (100 µL, 10 M) followed by the extraction with dichloromethane (600 µL). The water layer was removed after centrifugation and the organic layer was dried with MgSO4. Conversion was determined by GC with an Agilent DB-1701 column. The enantiomeric excess of the amine product was determined after derivatization to acetamido. Derivatization of the samples was performed by adding 4-dimethylaminopyridine in acetic anhydride (40 µL of stock solution 50 mg mL−1). The samples were shaken in an incubator at RT for 30 minutes. Afterwards water (150 µL) was added and the samples were shaken for additional 30 minutes. After centrifugation, the organic layer was dried with MgSO4. Enantiomeric excess was determined by GC with a Variant Chiracel DEX-CB column. Details on the GC analysis and methods are reported in the ESI paragraph S14 and S15.†
General optimized procedure for the orthogonal biocatalytic asymmetric amination of racemic alcohols on analytical scale
The procedure was the same as reported above with two differences. Strep-II-tagged TeS-ADH I86A W110A (45 µM) or a combination of Strep-II-tagged TeS-ADH W110A (23 µM) plus Lb-ADH (23 µM) were used. Racemic alcohols (20 mM) were used as substrates.
Preparative scale procedure for the orthogonal biocatalytic amination of 1m to give (R)-3m
NAD+ and NADP+ (final concentration 0.5 mM each) were dissolved in ammonium formate buffer (87 mL, 1 M, pH 8.5) in a 250 mL bottle with cap. Enzymes Strep-II-tagged TeS-ADH I86A W110A (45 µM), YcnD (10 µM), Ch1-AmDH (126 µM), Cb-FDH (20 µM) and catalase (0.2 μM) were added. Finally, the substrate rac-1m (50 mg) was added. The reaction mixture was incubated in an orbital shaker (30 °C, 170 rpm) and the progress of the reaction was monitored by TLC and GC. When quantitative conversion was achieved (ca. 12 h), the reaction mixture was acidified to pH 2–4 via addition of HCl (1 M) while cooling in an ice bath. The water layer was washed with methyl tert-butyl ether (15 mL) to remove any possible remaining alcohol starting material and ketone intermediate. The pH of the water phase was increased to basic pH via KOH (10 M) while cooling in an ice bath. The water layer was extracted with methyl tert-butyl ether (2 × 15 mL). The organic fractions containing the amine product were combined and dried over MgSO4. After filtration and evaporation of the solvent, the product was obtained in pure form (45 mg, 90% yield). Column chromatography was not required. The authenticity of the product was confirmed by 1H-NMR (ESI, paragraph S13†). 1H NMR (400 MHz, CDCl3, δ ppm): 2.87 (m, 1H, CH), 1.48 (s 2H, NH2), 1.28 (m, 8H, CH2), 1.05 (d, J = 6.9 Hz, 3H, CH3), 0.89 (t, J = 6.9 Hz, 3H, CH3).
In silico design of the 3D-structural models of WT TeS-ADH, TeS-ADH W110A, TeS-ADH I86A and TeS-ADH I86A W110A
The crystal structure of the thermophilic alcohol dehydrogenase from Thermoanaerobacter brockii (Tb-ADH, PDB code 1YKF)69 was used as the structural scaffold for the design of the model of the WT TeS-ADH. This scaffold was selected after a careful comparison between the sequence of the WT TeS-ADH and all the available 3D-structures and related sequences of the ADHs in the protein data bank (PDB). The sequences of Tb-ADH and WT-TeS-ADH differ only for 3 amino acids positions, namely: R91W, R313P, and R325Q (for sequence alignment, ESI paragraph 12†). These 3 mutations were induced in silico in order to generate the model of the WT TeS-ADH.
All in silico modifications were done with the Yasara software,73 utilizing the AMBER 03 force field.74 The protonation state of all atoms was automatically adjusted, with the exception of those atoms involved in the hydride transfer between co-factor and the substrate. The protonation state of the latter atoms was adjusted manually accordingly to their biochemical properties. Each mutation in the template structure was introduced singularly, one after the other. Furthermore, a three-step energy minimization procedure was executed for each mutation. The steps were as follows: (i) step one – only the mutated amino acid residue was energetically minimized; (ii) step two – the mutated residue plus all the residues located within a radius of 6 Å from the mutated residue were subjected to energy minimization; (iii) step three – the complete enzyme scaffold was subjected to energy minimization. The use of this protocol of energy minimization assures a gradual adjustment of the complete structure to the new mutation, thus avoiding the production of undesired secondary structure deformations.
Utilizing the same protocol as described above, the TeS-ADH variants (i.e. W110A, I86A, and I86A W110A) were generated from the model of WT TeS-ADH. We point out that the model of WT TeS-ADH was created containing acetone as substrate and Zn2+ as the metal cofactor in its active site. The position of the substrate and of Zn2+ was obtained from another available 3D structure of thermophilic alcohol dehydrogenase from Thermoanaerobacter brockii (PDB code 1BXZ) that also contains Zn2+ ion and 2-butanol as substrate in its active site.75 This structural information was utilized to introduce the substrate acetone in its reactive conformation into the active site of all the models of TeS-ADHs (WT, variant W110A, variant I86A and variant I86A W110A). The standardized reactive conformation of the ketone substrate was stabilized considering that the oxygen atom of the carbonyl group is in direct interaction with the Zn2+ ion (1.5–2.5 Å C═⋯Zn2+) and the hydride atom of the cofactor NADPH is in direct interaction with the C atom of the same carbonyl group (1.5–3.5 Å O═⋯H). The Zn2+ ion is kept in place by the interaction with the residues C37, H60, E60, and D150. For each TeS-ADH variant, a set of models was generated through the in situ introduction of each selected substrates (2b, 2p, 2u, 2g, and 2n). Each substrate was introduced starting from the spatial position of the acetone. Notably, pairs of enzyme plus substrate models were generated, wherein the substrate is either in the pro-S or pro-R conformation. For each enzyme plus substrate model, the three-step energy minimization protocol was applied as explained before. After energy minimization, aiming at further structural relaxation, a short molecular dynamic simulation (50 ps) was executed. Each final relaxed structure was evaluated using the Autodock Vina scoring function in order to assess its binding energy at the respective reactive conformation. Thus, two main parameters for the scoring function were used to rank the enzymatic complexes: (i) the distance between the hydride atom of the cofactor NADPH and the C atom of carbonyl group of the ketone substrate; (ii) the binding energy of the substrate in its reactive conformation. The following scoring function was utilized:
where is the relative score of enzymatic preference of the pro-S substrate over the pro-R one and are the distance H–C of the pro-S and pro-R substrate respectively; and are the binding energy of the pro-S and pro-R substrate respectively; rref is the reference H–C distance that was considered as the sum of the van der Waal radius of H and C (2.9 Å); R is the gas constant and T is the temperature of the simulations (298.15 K). The weighting parameter a and b were arbitrarily chosen to be a equal to 0.75 and b equal to 0.25.
All models herein were generated as homotetramers and each monomer possesses its own active site. Furthermore, each monomer was considered as an independent simulation from each other; thus each result is reported as the average observation over four active sites.
Conclusions
In this work, we present an atom-efficient artificial biocatalytic network that enables the one-pot stereoselective and up to quantitative conversion of alcohol functionalities into amines. The overall artificial biocatalytic network proceeds in a concurrent fashion without compartmentalization. This network constitutes therefore an elegant example of orthogonal tandem catalysis.72 We point out that our biocatalytic network exploits a high level of orthogonality because the biocatalysts possess – at the same time – high specificity for the type of compound and coenzyme without detectable cross-reactivity. On the other hand, the biocatalytic network is substrate promiscuous, as it possesses the capability of converting a range of structurally diverse alcohols.
The advantage of the herein described orthogonal biocatalytic network compared with the biocatalytic hydrogen-borrowing amination,18 previously published by our group, is the more favorable thermodynamic equilibrium (Scheme 1) that allows for obtaining >99% conversion for the amination of secondary alcohols. In fact, the estimated K′eq for the amination of secondary alcohol via the orthogonal network is 1.48 × 1041. In comparison the K′eq for the hydrogen-borrowing amination on the same substrate is 1.27.
Furthermore, the kinetics of the amination is also improved as the reaction times are reduced from 48 h (hydrogen-borrowing process) to 6 h (orthogonal network) as shown in Fig. 3. The orthogonal biocatalytic network shows an elevated atom efficiency as the reaction buffer, ammonium formate, is both the aminating agent and the source of reducing equivalents in the second step. Overall, the amination of 1 mole of alcohol requires 1 mole of ammonium formate and generates 1 mole of carbonate. Dioxygen is the only other reagent. The only drawback of herein described orthogonal biocatalytic network compared with the biocatalytic hydrogen-borrowing amination is the requirement for two additional enzymes (YcnD and FDH) for the recycling of the nicotinamide coenzymes. Nevertheless, this drawback can be alleviated if the enzymes involved in the cascade are coexpressed altogether in E. coli as host organism and whole cells are used as catalysts. Recent publications have shown that it is possible to produce up to eight heterologous enzymes in E. coli with a perfect balance between enzyme expressions and activities.76,77
The applicability of the orthogonal biocatalytic network requires a non-enantioselective ADH in the oxidative step. The I86A W110A variant of the TeS-ADH was suitable for the oxidation of a number of racemic alcohols. The effect of the mutations were studied with a rather straightforward in silico approach. However, an excellent agreement between the models obtained and the experimental data was observed. The in silico approach used in the present work pre-assumes that all the substrates would have the same probability to reach the active site independently from the pro-S or pro-R conformation. However, some enzymes are capable of pre-orienting and redirecting the substrate into the most accepted conformation at the entrance tunnel of the enzyme. Furthermore, a less static approach (e.g. the inclusion of longer and repetitive molecular dynamics simulations) might allow for a more detailed understanding of the catalytic properties of the variants. Besides, the engineering of amine dehydrogenases for the conversion of ketones into S-configured amines will complement the scope of this artificial biocatalytic network.
In summary, this work represents a valuable addition to the repertoire of the catalytic methods for the stereoselective amination of alcohols and it will open new perspective in the field of protein engineering of oxidoreductases.
Supplementary Material
† Electronic supplementary information (ESI) available: Site directed mutagenesis, expression and purification conditions for the recombinant enzymes, analytical methods as well as GC chromatograms and calculations of Gibbs free energies. See DOI: 10.1039/c7ob01927k
Acknowledgements
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 638271, BioSusAmin). Dutch funding from the NWO Sector Plan for Physics and Chemistry is also acknowledged.
We thank Prof. Peter Macheroux for kindly donating the plasmid for the expression of YcnD.
Footnotes
Conflicts of interest
The authors declare to have no competing interests, or other interests that might be perceived to influence the results and/or discussion reported in this article.
Tanja Knaus: 0000-0001-9942-9226
Luca Cariati: 0000-0002-2671-949X
Marcelo F. Masman: 0000-0002-9849-3901
Francesco G. Mutti: 0000-0002-6771-5102
Notes and references
- 1.Wittcoff HA, Rueben BG, Plotkin JS. Industrial Organic Chemicals. 2nd edn. Wiley-Interscience; New York: 2004. [Google Scholar]
- 2.Nugent TC, editor. Chiral amine synthesis: Methods, Developments and Applications. Wiley-VCH; Weinheim: 2010. [Google Scholar]
- 3.Au SK, Groover J, Feske BD, Bommarius AS. Organic Synthesis Using Biocatalysis. 1st edn. Elsevier; 2015. pp. 187–212. [Google Scholar]
- 4.Fabiano E, Golding BT, Sadeghi MM. Synthesis. 1987:190–192. [Google Scholar]
- 5.Swamy KC, Kumar NN, Balaraman E, Kumar KV. Chem Rev. 2009;109:2551–2651. doi: 10.1021/cr800278z. [DOI] [PubMed] [Google Scholar]
- 6.Iula DM. In: Name Reactions for Functional Group Transformations. Li JJ, Corey EJ, editors. John Wiley & Sons, Inc.; 2007. pp. 129–151. [Google Scholar]
- 7.Bähn S, Imm S, Neubert L, Zhang M, Neumann H, Beller M. ChemCatChem. 2011;3:1853–1864. [Google Scholar]
- 8.Grigg R, Mitchell TRB, Sutthivaiyakit S, Tongpenyai N. J Chem Soc, Chem Commun. 1981:611. [Google Scholar]
- 9.Gunanathan C, Milstein D. Angew Chem, Int Ed. 2008;47:8661–8664. doi: 10.1002/anie.200803229. [DOI] [PubMed] [Google Scholar]
- 10.Imm S, Bahn S, Neubert L, Neumann H, Beller M. Angew Chem, Int Ed. 2010;49:8126–8129. doi: 10.1002/anie.201002576. [DOI] [PubMed] [Google Scholar]
- 11.Pingen D, Muller C, Vogt D. Angew Chem, Int Ed. 2010;49:8130–8133. doi: 10.1002/anie.201002583. [DOI] [PubMed] [Google Scholar]
- 12.Pingen D, Diebolt O, Vogt D. ChemCatChem. 2013;5:2905–2912. [Google Scholar]
- 13.Saidi O, Blacker AJ, Farah MM, Marsden SP, Williams JM. Angew Chem, Int Ed. 2009;48:7375–7378. doi: 10.1002/anie.200904028. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Lim CS, Sim DS, Pan HJ, Zhao Y. Angew Chem, Int Ed. 2014;53:1399–1403. doi: 10.1002/anie.201307789. [DOI] [PubMed] [Google Scholar]
- 15.Blank B, Michlik S, Kempe R. Chem – Eur J. 2009;15:3790–3799. doi: 10.1002/chem.200802318. [DOI] [PubMed] [Google Scholar]
- 16.Oldenhuis NJ, Dong VM, Guan Z. J Am Chem Soc. 2014;136:12548–12551. doi: 10.1021/ja5058482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leonard J, Blacker AJ, Marsden SP, Jones MF, Mulholland KR, Newton R. Org Process Res Dev. 2015;19:1400–1410. [Google Scholar]
- 18.Mutti FG, Knaus T, Scrutton NS, Breuer M, Turner NJ. Science. 2015;349:1525–1529. doi: 10.1126/science.aac9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen F-F, Liu Y-Y, Zheng G-W, Xu J-H. ChemCatChem. 2015;7:3838–3841. [Google Scholar]
- 20.Wang JB, Reetz MT. Nat Chem. 2015;7:948–949. doi: 10.1038/nchem.2408. [DOI] [PubMed] [Google Scholar]
- 21.Hollmann F. In: Science of Synthesis, Biocatalysis in Organic Synthesis 3. Faber k, Fessner W-D, Turner NJ., editors. Georg Thieme Verlag KG; Stuttgart (Germany): 2015. pp. 115–138. [Google Scholar]
- 22.Moody TS, Mix S, Brown G, Beecher D. In: Science of Synthesis, Biocatalysis in Organic Synthesis 2. Faber K, Fessner W-D, Turner NJ, editors. Georg Thieme Verlag KG; Stuttgart (Germany): 2015. pp. 421–458. [Google Scholar]
- 23.Bommarius AS, Au SK. In: Science of Synthesis, Biocatalysis in Organic Synthesis 2. Faber K, Fessner W-D, Turner NJ, editors. Georg Thieme Verlag KG; Stuttgart (Germany): 2015. pp. 335–357. [Google Scholar]
- 24.Knaus T, Böhmer W, Mutti FG. Green Chem. 2017;19:453–463. doi: 10.1039/C6GC01987K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye LJ, Toh HH, Yang Y, Adams JP, Snajdrova R, Li Z. ACS Catal. 2015;5:1119–1122. [Google Scholar]
- 26.Au SK, Bommarius BR, Bommarius AS. ACS Catal. 2014;4:4021–4026. [Google Scholar]
- 27.Abrahamson MJ, Vazquez-Figueroa E, Woodall NB, Moore JC, Bommarius AS. Angew Chem, Int Ed. 2012;51:3969–3972. doi: 10.1002/anie.201107813. [DOI] [PubMed] [Google Scholar]
- 28.Bommarius BR, Schürmann M, Bommarius AS. Chem Commun. 2014;50:14953–14955. doi: 10.1039/c4cc06527a. [DOI] [PubMed] [Google Scholar]
- 29.Reaction conditions: 20 mM substrate, 2 M of total NH4+/NH3, 1 mM NAD+.
- 30.Sattler JH, Fuchs M, Tauber K, Mutti FG, Faber K, Pfeffer J, Haas T, Kroutil W. Angew Chem, Int Ed. 2012;51:9156–9159. doi: 10.1002/anie.201204683. [DOI] [PubMed] [Google Scholar]
- 31.Tauber K, Fuchs M, Sattler JH, Pitzer J, Pressnitz D, Koszelewski D, Faber K, Pfeffer J, Haas T, Kroutil W. Chem – Eur J. 2013;19:4030–4035. doi: 10.1002/chem.201202666. [DOI] [PubMed] [Google Scholar]
- 32.Lerchner A, Achatz S, Rausch C, Haas T, Skerra A. ChemCatChem. 2013;5:3374–3383. [Google Scholar]
- 33.Palacio CM, Crismaru CG, Bartsch S, Navickas V, Ditrich K, Breuer M, Abu R, Woodley JM, Baldenius K, Wu B, Janssen DB. Biotechnol Bioeng. 2016;113:1853–1861. doi: 10.1002/bit.25954. [DOI] [PubMed] [Google Scholar]
- 34.Klatte S, Wendisch VF. Bioorg Med Chem. 2014;22:5578–5585. doi: 10.1016/j.bmc.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 35.Morokutti A, Lyskowski A, Sollner S, Pointner E, Fitzpatrick TB, Kratky C, Gruber K, Macheroux P. Biochemistry. 2005;44:13724–13733. doi: 10.1021/bi0510835. [DOI] [PubMed] [Google Scholar]
- 36.Schutte H, Flossdorf J, Sahm H, Kula M-R. Eur J Biochem. 1976;62:151–160. doi: 10.1111/j.1432-1033.1976.tb10108.x. [DOI] [PubMed] [Google Scholar]
- 37.Wong CH, Zimmerman SC. Chem Commun. 2013;49:1679–1695. doi: 10.1039/c2cc37316e. [DOI] [PubMed] [Google Scholar]
- 38.Voss CV, Gruber CC, Faber K, Knaus T, Macheroux P, Kroutil W. J Am Chem Soc. 2008;130:13969–13972. doi: 10.1021/ja804816a. [DOI] [PubMed] [Google Scholar]
- 39.Schrittwieser JH, Velikogne S, Hall M, Kroutil W. Chem Rev. 2017 doi: 10.1021/acs.chemrev.7b00033. [DOI] [PubMed] [Google Scholar]
- 40.Monti D, Ferrandi EE, Zanellato I, Hua L, Polentini F, Carrea G, Riva S. Adv Synth Catal. 2009;351:1303–1311. [Google Scholar]
- 41.Adam W, Lazarus M, Saha-Möller CR, Schreier P. Tetrahedron: Asymmetry. 1998;9:351–355. [Google Scholar]
- 42.Xue YP, Zeng H, Jin XL, Liu ZQ, Zheng YG. Microb Cell Fact. 2016;15:162. doi: 10.1186/s12934-016-0560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Díaz-Rodríguez A, Ríos-Lombardía N, Sattler JH, Lavandera I, Gotor-Fernández V, Kroutil W, Gotor V. Catal Sci Technol. 2015;5:1443–1446. [Google Scholar]
- 44.Fuchs M, Tauber K, Sattler J, Lechner H, Pfeffer J, Kroutil W, Faber K. RSC Adv. 2012;2:6262. [Google Scholar]
- 45.Pickl M, Fuchs M, Glueck SM, Faber K. ChemCatChem. 2015;7:3121–3124. doi: 10.1002/cctc.201500589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martínez-Montero L, Gotor V, Gotor-Fernández V, Lavandera I. Green Chem. 2017;19:474–480. [Google Scholar]
- 47.ΔrG° is the change in Gibbs free energy due to a chemical reaction in standard conditions (T 298.15 K, P 1 bar) and without accounting for pH and ionic strength. ΔrG′° represents the change in Gibbs free energy due to a chemical reaction in standard conditions considering a particular pH and ionic strength. In this case, we set pH 8.5 and ionic strenght 1 M.
- 48.Heiss C, Phillips RS. J Chem Soc Perkin Trans 1. 2000:2821–2825. [Google Scholar]
- 49.Zheng C, Pham VT, Phillips RS. Catal Today. 1994;22:607–620. [Google Scholar]
- 50.Zheng C, Pham VT, Phillips RS. Bioorg Med Chem Lett. 1992;2:619–622. [Google Scholar]
- 51.Burdette D, Zeikus JG. Biochem J. 1994;302:163–170. doi: 10.1042/bj3020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burdette DS, Vieille C, Zeikus JG. Biochem J. 1996;316:115–122. doi: 10.1042/bj3160115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Musa MM, Ziegelmann-Fjeld KI, Vieille C, Zeikus JG, Phillips RS. Angew Chem, Int Ed. 2007;46:3091–3094. doi: 10.1002/anie.200604615. [DOI] [PubMed] [Google Scholar]
- 54.Musa MM, Ziegelmann-Fjeld KI, Vieille C, Zeikus JG, Phillips RS. J Org Chem. 2007;72:30–34. doi: 10.1021/jo0616097. [DOI] [PubMed] [Google Scholar]
- 55.Ziegelmann-Fjeld KI, Musa MM, Phillips RS, Zeikus JG, Vieille C. Protein Eng, Des Sel. 2007;20:47–55. doi: 10.1093/protein/gzl052. [DOI] [PubMed] [Google Scholar]
- 56.Musa MM, Lott N, Laivenieks M, Watanabe L, Vieille C, Phillips RS. ChemCatChem. 2009;1:89–93. [Google Scholar]
- 57.Patel JM, Musa MM, Rodriguez L, Sutton DA, Popik VV, Phillips RS. Org Biomol Chem. 2014;12:5905–5910. doi: 10.1039/c4ob00794h. [DOI] [PubMed] [Google Scholar]
- 58.Karume I, Takahashi M, Hamdan SM, Musa MM. ChemCatChem. 2016;8:1459–1463. [Google Scholar]
- 59.Musa MM, Patel JM, Nealon CM, Kim CS, Phillips RS, Karume I. J Mol Catal B: Enzym. 2015;115:155–159. [Google Scholar]
- 60.Nealon CM, Welsh TP, Kim CS, Phillips RS. Arch Biochem Biophys. 2016;606:151–156. doi: 10.1016/j.abb.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 61.Karume I, Musa MM, Bsharat O, Takahashi M, Hamdan SM, El Ali B. RSC Adv. 2016;6:96616–96622. [Google Scholar]
- 62.Musa MM, Phillips RS, Laivenieks M, Vieille C, Takahashi M, Hamdan SM. Org Biomol Chem. 2013;11:2911–2915. doi: 10.1039/c3ob27415b. [DOI] [PubMed] [Google Scholar]
- 63.Schlieben NH, Niefind K, Muller J, Riebel B, Hummel W, Schomburg D. J Mol Biol. 2005;349:801–813. doi: 10.1016/j.jmb.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 64.Rodriguez C, Borzecka W, Sattler JH, Kroutil W, Lavandera I, Gotor V. Org Biomol Chem. 2014;12:673–681. doi: 10.1039/c3ob42057d. [DOI] [PubMed] [Google Scholar]
- 65.Cuetos A, Rioz-Martínez A, Bisogno FR, Grischek B, Lavandera I, de Gonzalo G, Kroutil W, Gotor V. Adv Synth Catal. 2012;354:1743–1749. [Google Scholar]
- 66.Naik HG, Yeniad B, Koning CE, Heise A. Org Biomol Chem. 2012;10:4961–4967. doi: 10.1039/c2ob06870b. [DOI] [PubMed] [Google Scholar]
- 67.Bisogno FR, Garcia-Urdiales E, Valdes H, Lavandera I, Kroutil W, Suarez D, Gotor V. Chem – Eur J. 2010;16:11012–11019. doi: 10.1002/chem.201001233. [DOI] [PubMed] [Google Scholar]
- 68.Bsharat O, Musa MM, Vieille C, Oladepo SA, Takahashi M, Hamdan SM. ChemCatChem. 2017;9:1487–1493. [Google Scholar]
- 69.Korkhin Y, Kalb AJ, Peretz M, Bogin O, Burstein Y, Frolow F. J Mol Biol. 1998;278:967–981. doi: 10.1006/jmbi.1998.1750. [DOI] [PubMed] [Google Scholar]
- 70.Sun Z, Lonsdale R, Ilie A, Li G, Zhou J, Reetz MT. ACS Catal. 2016;6:1598–1605. [Google Scholar]
- 71.Abrahamson MJ, Wong JW, Bommarius AS. Adv Synth Catal. 2013;355:1780–1786. [Google Scholar]
- 72.Lohr TL, Marks TJ. Nat Chem. 2015;7:477–482. doi: 10.1038/nchem.2262. [DOI] [PubMed] [Google Scholar]
- 73.Krieger E, Koraimann G, Vriend G. Proteins: Struct, Funct, Genet. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- 74.Oostenbrink C, Villa A, Mark AE, van Gunsteren WF. J Comput Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 75.Li C, Heatwole J, Soelaiman S, Shoham M. Proteins: Struct, Funct, Genet. 1999;37:619–627. doi: 10.1002/(sici)1097-0134(19991201)37:4<619::aid-prot12>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 76.Zhou Y, Wu S, Li Z. Angew Chem, Int Ed. 2016;55:11647–11650. doi: 10.1002/anie.201606235. [DOI] [PubMed] [Google Scholar]
- 77.Wu S, Zhou Y, Wang T, Too HP, Wang DI, Li Z. Nat Commun. 2016;7:11917. doi: 10.1038/ncomms11917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.