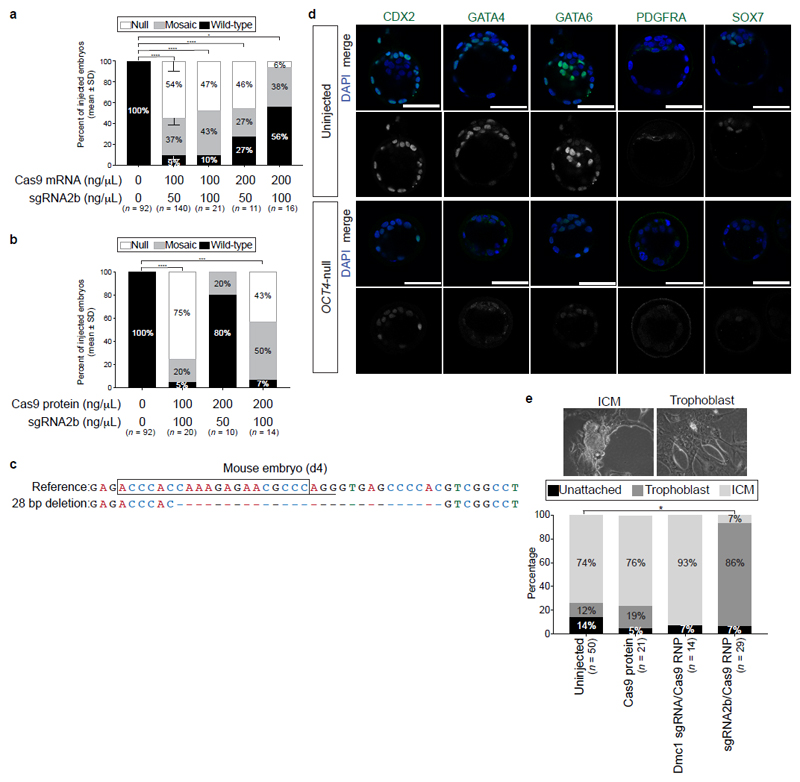
Extended Data Figure 5. Assessing a range of Cas9 and sgRNA combinations for microinjection into mouse pronuclear zygotes.
a, b, Additional conditions were tested in mouse embryos microinjected with the sgRNA2b either with Cas9 mRNA (a) or as a complex with the Cas9 protein (b) at the ratios indicated. Quantification was performed on the proportion of mouse embryos at the blastocyst stage that are phenotypically null (loss of OCT4 and SOX17 protein expression), mosaic or heterozygous (partial OCT4 and/or SOX17 expression) or uninjected (strong OCT4 and SOX17 expression). Data are mean ± s.d. from three independent experiments. Comparisons were made between the percentage of OCT4-null embryos observed versus wild-type uninjected control embryos. Chi-squared test. *P < 0.05; ***P < 0.001; ****P < 0.0001. c, The types of indel mutations detected in mouse embryos microinjected with the sgRNA2b–Cas9 complex. The sgRNA sequence is boxed and the NGG PAM site underlined. Dash, deletion position. d, Further characterization of mouse embryos microinjected with sgRNA2b–Cas9 compared to uninjected control blastocysts. Immunofluorescence analysis for markers of the trophectoderm (CDX2) or primitive endoderm (GATA4, GATA6, PDGFRA and SOX7) lineages together with DAPI nuclear staining. Confocal z-section. Scale bar, 100 μm. e, Quantification of blastocyst inner cell mass (ICM) or trophoblast outgrowths in mouse embryonic stem cell derivation conditions. Uninjected, Cas9-injected or Cas9 plus Dmc1 sgRNA-injected cells (targeting a gene not essential for preimplantation development) were used as controls. Comparisons were made between the percentage of ICM outgrowths observed in blastocysts that developed following sgRNA2b–Cas9 microinjection. Two-tailed t-test. *P < 0.05.