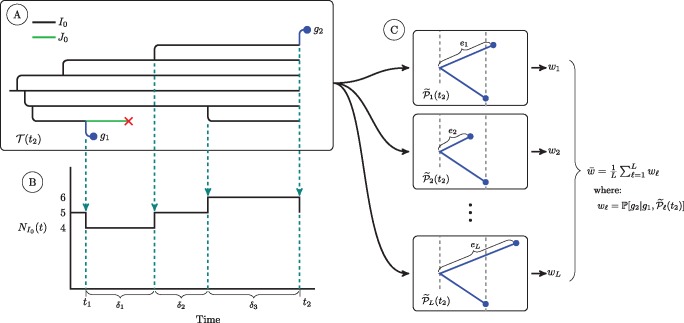
Fig. 10.
A schematic of quantities used in calculation of the conditional density of a diagnosis and the conditional probability of a genetic sequence. At (A) we show a simulated transmission tree. For simplicity, this tree only has individuals of class I0 and class J0. Dashed arrows fall from events in the transmission tree that change the count of I0 individuals in the population. At (B) we show a plot of the trajectory of the I0 class. This plot shows the quantities we use to calculate the cumulative hazard of diagnosis for the I0 class, , over an interval of time from t1 to t2. We first subdivide the time interval into R subintervals over which the number of I0 individuals is constant (indicated with dashed lines). We let the number of I0 individuals in the rth subinterval be . The cumulative hazard of diagnosis is then: . The cumulative hazards of diagnosis for the other two classes of undiagnosed individuals are computed in the same fashion. At (C) we show the set of L subtrees of the phylogeny that we use to numerically estimate the conditional probability of sequence g2 under our relaxed clock model. The subtree is constructed by augmenting with a new edge with length drawn from a Gamma distribution parameterized as described in the text. For each of these L subtrees we use the peeling algorithm to compute , the conditional probability of observing sequence g2 given sequence g1 and the structure of . The average of these conditional probabilities is a numerical estimate of the conditional probability of g2 under our relaxed clock model. For simplicity, here we do not show the case in which the edge length of g2 splits an existing edge; this case requires a beta bridge to apportion the length of the split edge so as to maintain Gamma distributed edge lengths. For this more complicated case, see figure 9.