Abstract
Numerous brain disorders are associated with ventriculomegaly; normal pressure hydrocephalus (NPH) is one example. NPH presents with dementia-like symptoms and is often misdiagnosed as Alzheimer’s due to its chronic nature and nonspecific presenting symptoms. However, unlike other forms of dementia NPH can be treated surgically with an over 80% success rate on appropriately selected patients. Accurate assessment of the ventricles, in particular its sub-compartments, is required to diagnose the condition. Existing segmentation algorithms fail to accurately identify the ventricles in patients with such extreme pathology. We present an improvement to a whole brain segmentation approach that accurately identifies the ventricles and parcellates them into four sub-compartments. Our work is a combination of patch-based tissue segmentation and multi-atlas registration-based labeling. We include a validation on NPH patients, demonstrating superior performance against state-of-the-art methods.
Keywords: Brain, MRI, Enlarged ventricles, Hydrocephalus
1 Introduction
The ventricular system of the human brain is made up of four cavities: the left and right lateral ventricles and the third and fourth ventricles. These cavities are connected via narrow channels, with the foramina of Monro connecting each of the lateral ventricles with the third ventricle and the cerebral aqueduct connecting the third and fourth ventricles. Each of these cavities contain choroid plexus, which is responsible for producing cerebrospinal fluid (CSF). In a healthy system, CSF is allowed to flow from the lateral ventricles into the third and then the fourth ventricle and subsequently into the central canal of the spinal cord and up into the subarachnoid space, before passing through the arachnoid villi into the venous sinuses.
Normal pressure hydrocephalus (NPH) is a disorder of the ventricular system caused by obstruction of the flow of CSF leading to the expansion of the cerebral ventricles and with symptoms including [1]: gait disturbance, urinary incontinence, and dementia. An example of a T1-weighted (T1-w) magnetization prepared rapid gradient echo (MPRAGE) of an NPH patient can be seen in Fig. 1(a). The expanded ventricular system presses against the surrounding structures causing the brain shape to become distorted and results in brain damage. NPH is routinely misdiagnosed as other forms of dementia, such as Parkinson’s disease or Alzheimer’s disease. However, unlike other forms of dementia, NPH is treatable and the associated symptoms can be reversed (to a certain extent) [10]. Treatment involves shunt surgery or endoscopic third ventriculostomy. However, diagnosing NPH patients is challenging using current methods, and the benefit of surgical intervention is sensitive to properly selected patients [14]. The chronically dilated ventricles are readily observed through magnetic resonance imaging (MRI), which when used in conjunction with a lumbar puncture and evaluation of the clinical response to removal of CSF can help to diagnose the condition. However, having accurate parcellation of the ventricular system into its sub-compartments would be of great clinical benefit to better characterize the pathology of NPH as well as to help in surgical planning, such that patients who will benefit from surgical treatments could be more robustly identified.
Fig. 1.
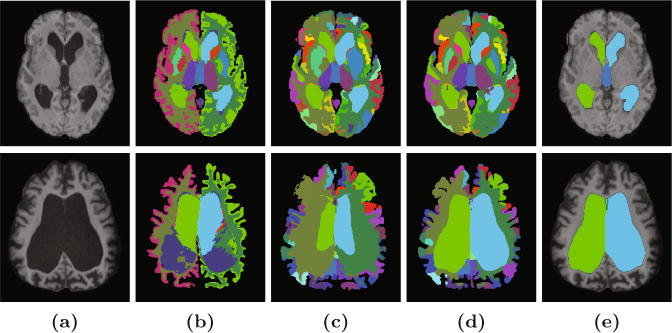
Shown in each row is the (a) T1-w MPRAGE of a NPH patient and the ventricle segmentation (green/cyan is the right/left lateral ventricle, blue is the 3rd ventricle) generated by (b) FreeSurfer [8], (c) MALPEM [9], (d) RUDOLPH, and (e) a manual delineation. The first row shows a NPH patient where all three algorithms performed well and the second row shows a more severe case where FreeSurfer and MALPEM have failed. The other colors show the rich tapestry of labels available in all three methods. (Color figure online)
Previous work on ventricle segmentation [5,13] has focused on the ventricular system as one component of the brain. Newer methods [9] that provide an improved ventricle segmentation rely, in part, on multi-atlas segmentation frameworks, which enable them to identify some of the components of the ventricles (right and left lateral, 3rd, 4th) based on the labels available within their atlases. These more recent methods, however, often fail to correctly identify the extents of the ventricles in pathological cases, see Fig. 1. This occurs chiefly because they depend on a registration between the atlas and subject, which in pathological cases is rarely optimum. To address this problem in our work, we incorporate a patch-based segmentation method [11,12] to provide a prior for a multi-atlas label fusion framework [9]. Our method, known as robust dictionary-learning and label propagation hybrid (RUDOLPH) [7], provides a parcellation of the entire brain, providing 138 brain labels (in the cerebellum and cerebrum) while performing accurate ventricular segmentation even with enlarged ventricles. We present a detailed evaluation of this method with respect to the four main cavities of the ventricular system of NPH patients (noting that RUDOLPH also provides a parcellation of the whole brain, examples of which can be seen in Fig. 1). In Sect. 2, we describe RUDOLPH. Section 3 includes our experiments comparing our approach and two state-of-the-art segmentation algorithms on our manual delineations. We conclude with a discussion of the presented work in Sect. 4.
2 Method
The proposed method integrates the subject specific sparse dictionary learning (S3DL) method [11,12] and the multi-atlas label propagation with expectation-maximization (MALPEM) method [9]. S3DL is a patch-based segmentation method that uses sparse dictionary learning to classify the human brain into seven structures (cerebellar and cerebral white matter; cortical, subcortical, and cerebellar gray matter; and ventricular and cortical CSF). MALPEM is a multi-atlas label fusion scheme, which we modify to incorporate the seven labels from S3DL, intelligently guiding the multi-atlas label fusion framework. MALPEM cannot, in general, segment the ventricles in moderate to severe cases of NPH patients, due to the pathology (see Fig. 1(b)).
We first process a subject’s MR image through S3DL. S3DL requires an atlas with its corresponding hard segmentation, and spatial priors depicting where the different tissues are expected to be located. The priors are computed using a simple blurring of the known atlas segmentation. S3DL adaptively modifies the subject priors to handle anatomical variability. The CSF labels from S3DL are incorporated within MALPEM, as described below.
We then register using SyN [2] 15 manually labeled atlases into the subject’s space, with each atlas made up of 138 cortical and subcortical labels from Neuromorphometrics1. This is similar to the first step of MALPEM, which uses 30 atlases. The output of this step is a probabilistic segmentation Π = {π1, …, πn}, where πi is a K-dimensional vector representing the K = 138 labels in the atlases, and n is the total number of voxels in the subject’s image. MALPEM provides two label correction schemes; the first based on intensity-refined posterior probabilities, and the second relaxes the probabilities, Π, to correct for misregistration of the atlases. The eight CSF labels within Neuromorphometrics are assumed to come from the Gaussian distribution (μCSF, σCSF) which is estimated based on Π. For each label k, (k = 1, …, K) we estimate (μk, σk) from the subject’s image intensities. Then Π is relaxed to ΠR using the distributions (μk, σk) as follows. At each voxel i, a fraction αik of prior πik is redistributed from label k to one of the eight CSF labels based on the spatial proximity of the label to the CSF label with the highest probability. Both of these conditions fail in NPH patients since at the boundary of severely dilated ventricles the closest CSF label is usually cortical CSF and not the desired ventricular CSF and the severe deformation of NPH patients means that spatial information from anatomical atlases is incorrect.
To address this, we use the segmentation from S3DL. Thus, we identify the appropriate CSF label kCSF as
| (1) |
where dk(i) is the distance from the voxel i to the nearest voxel with the current label k, and and are the CSF labels of MALPEM and S3DL, respectively. We follow the MALPEM framework and compute the relaxation fraction, αik, based on the probability that the voxel x comes from either the intensity distribution estimated by label k or from one of the CSF distributions estimated by ,
| (2) |
The relaxed prior ΠR is computed as
| (3) |
ΠR is then updated through an expectation-maximization framework [15], with smoothness of the final segmentation maintained through a Markov Random Field [17], which is the same as in the MALPEM framework.
3 Results
Our data was acquired on a Siemens 3 T scanner using a T1-w MPRAGE with TR = 10.3 ms, TE = 6 ms, and 0.82 × 0.82 × 1.17 mm3 voxel size. We processed a total of 45 NPH patients that were broadly classified based on the severity of their ventricular expansion into mild, moderate, and severe cases. All 45 NPH patients had their ventricular system manually delineated. This was done by identifying the anatomical structure of the ventricles, which required 3–4 h per patient. These were reviewed by separate experts in neuroanatomy, with possible correction or return to the delineator for correction. For 18 of the 45 NPH patients, once a ventricular system mask was agreed, the components of right and left lateral ventricles, both foramina of Monro, third ventricle, cerebral aqueduct, and fourth ventricle were identified. This parcellation of the ventricular system took another hour per patient to complete. The cerebral aqueduct and the foramina of Monro are not included within our validation as there do not currently exist such detailed anatomical atlases of the ventricular system in use elsewhere. Thus for validation purposes, the foramina of Monro is included with the corresponding lateral ventricle, and the cerebral aqueduct with the fourth ventricle, making the labeling comparable with Neuromorphometrics.
We processed the 45 NPH patients using RUDOLPH and two state-of-the-art whole brain segmentation methods: FreeSurfer (Version 5.3.0) [8] and MALPEM [9]. We ran FreeSurfer with the - bigventricles flag. We used an in-house [3,4] approach to skull strip the data, as we have found that this improves the performance of both FreeSurfer and MALPEM on our NPH cohort. The volumes generated by FreeSurfer, MALPEM, and RUDOLPH for the entire ventricular system are shown in the top row of Fig. 2; the patients are ordered based on the volume of the manual delineations which is also shown in the figure. The NPH patient shown in the top row of Fig. 1 corresponds to Patient ID #39 in Fig. 2, the bottom row of Fig. 1 corresponds to Patient ID #43. We computed the Dice coefficient [6] for these 45 NPH patients on the entire ventricle system, the results are reported in Table 1. A paired two-sided Wilcoxon Signed-Rank Test [16], without a correction for multiple comparisons, comparing FreeSurfer to MALPEM on the entire ventricle system yielded significant differences with a p-value <0.001. We also obtained a similar p-value (<0.001) when comparing MALPEM to RUDOLPH on the entire ventricle system. These Dice coefficients are shown in the bottom row of Fig. 2, ordered by the volume of the manual masks. We also computed the Dice coefficient for each of the automatically labeled ventricular cavities with the corresponding manual delineation; see Table 1 and Fig. 3. RUDOLPH produces more accurate segmentation of the third ventricle and the left and right lateral ventricles than both MALPEM and FreeSurfer; these results also reach statistical significance. The fourth ventricle is most accurately segmented by MALPEM, however it is not statistically significantly better than RUDOLPH (see Table 2).
Fig. 2.
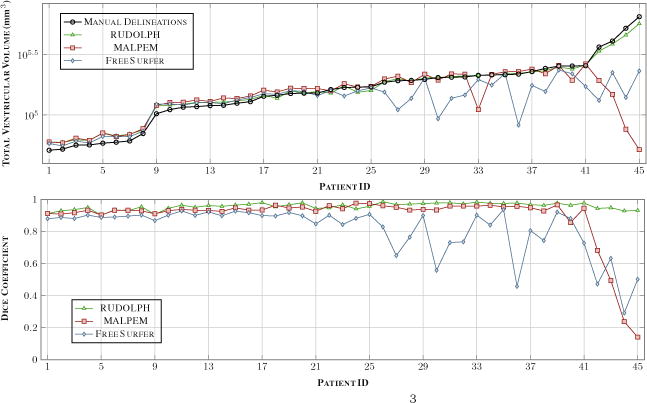
The top row shows the mean volume in mm3 for the manual masks and the three methods over the 45 NPH patients, ordered based on the volume of the manual masks. The y-axis uses a log scale to help differentiate the volumes of the different methods across the whole range of volumes. The bottom row shows the Dice coefficient over the same 45 NPH patients with the same ordering. The NPH patient shown in the top row of Fig. 1 has Patient ID #39 and the bottom row corresponds to Patient ID #43.
Table 1.
The mean Dice coefficient (and standard deviation) over our population of NPH patients measuring similarity between manual labels and automatically generated labels from the three methods. For 45 NPH patients we compare the entire (Entire) ventricle system. For 18 of those 45 we compare four ventricular cavities, third ventricle (3rd), fourth ventricle (4th), right lateral ventricle (RLV), and left lateral ventricle (LLV).
| FreeSurfer | MALPEM | RUDOLPH | |
|---|---|---|---|
| Entire | 0.815 (±0.150) | 0.890 (±0.172) | 0.957 (±0.021) |
| 3rd | 0.775 (±0.091) | 0.780 (±0.140) | 0.869 (±0.065) |
| 4th | 0.656 (±0.156) | 0.720 (±0.152) | 0.694 (±0.205) |
| RLV | 0.799 (±0.181) | 0.810 (±0.278) | 0.956 (±0.022) |
| LLV | 0.803 (±0.192) | 0.825 (±0.262) | 0.959 (±0.018) |
Fig. 3.
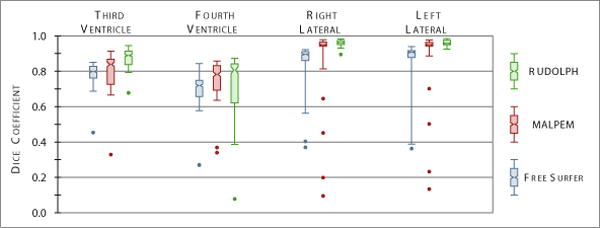
Box plots of the Dice coefficient with respect to our manual masks over 18 NPH patients comparing the automatically generated labels from the three methods for four ventricular cavities: third ventricle, fourth ventricle, right lateral ventricle, and left lateral ventricle.
Table 2.
p-values for a paired two-sided Wilcoxon Signed-Rank Test [16], without a correction for multiple comparisons, between the two methods listed for the noted ventricle cavity. This is across the 18 patients that are also compared to manual masks and presented in Fig. 3. The key for the ventricular cavities is: third ventricle – 3rd; fourth ventricle – 4th; right lateral ventricle – RLV; and left lateral ventricle – LLV.
| Comparison | 3rd | 4th | RLV | LLV |
|---|---|---|---|---|
| FreeSurfer vs. MALPEM | 0.6397 | 0.0007 | 0.2837 | 0.0987 |
| FreeSurfer vs. RUDOLPH | 0.0007 | 0.1187 | 0.0000 | 0.0000 |
| MALPEM vs. RUDOLPH | 0.0007 | 0.2462 | 0.0023 | 0.0016 |
4 Discussion and Conclusions
We have presented a method for whole brain segmentation that provides a robust segmentation of the ventricular system in patients with severely enlarged ventricles. We have shown that the approach is more robust on the ventricles than either FreeSurfer or MALPEM across 45 NPH patients; it consistently tracks the volume generated by the manual delineation of the ventricles better than either method as shown in the top row of Fig. 2. In particular, we note that both the FreeSurfer and MALPEM estimates of the ventricular CSF volume become more erratic as the volume increases. This is particularly troubling as the ventricles do naturally increase in size through natural brain atrophy over the time course of healthy patients. This study would call into question the validity of using these methods in a fully automated fashion without some quality assurance review of the results. We also note that FreeSurfer appears to level off and be unable to provide ventricular volumes above a certain level, which may be a limitation of the approach. MALPEM also exhibits instability in its results as the ventricular volume increases (see Fig. 2), however these do not always seem to be tied to the volume of the ventricles. As noted earlier the MALPEM estimates of the ventricular volume for Patient ID #39 are reasonable. Yet Patient ID #40 has a similar volume to Patient ID #39 and MALPEM performs poorly; whereas Patient ID #36 has less volume and MALPEM essentially fails. The results suggest that these patients may have significantly differently shaped structures which is leading to the failure of MALPEM. Our initial review of these results suggest that misregistrations within the multi-atlas phase of MALPEM may be the cause of these problems; which reinforces our belief that our enhancements to MALPEM are appropriate fixes for pathology cases. We also demonstrated that our approach can more accurately estimate the ventricular cavities of the lateral ventricles and the third ventricle (see Table 1).
Future work includes creating manual delineations on a larger cohort of patients—in particular patients suffering from ventriculomegaly by other causes. We also plan to further refine the parcellation of the ventricles to include the subchambers—anterior, occipital, and temporal horns of the lateral ventricles. A future goal will be correlating the volumetrics of these structures with surgical outcomes for NPH patients.
Acknowledgments
This work was supported by the NIH/NINDS under grant R21-NS096497. Support was also provided by the National Multiple Sclerosis Society grant RG-1507-05243 and the Dept. of Defense Center for Neuroscience and Regenerative Medicine.
Footnotes
References
- 1.Adams RD, et al. Symptomatic occult hydrocephalus with normal cerebrospinal-fluid pressure - a treatable syndrome. New Eng J Med. 1965;273(3):117–126. doi: 10.1056/NEJM196507152730301. [DOI] [PubMed] [Google Scholar]
- 2.Avants BB, et al. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carass A, et al. 4th International Symposium on Biomedical Imaging (ISBI 2007) IEEE; 2007. A joint registration and segmentation approach to skull stripping; pp. 656–659. [Google Scholar]
- 4.Carass A, et al. Simple paradigm for extra-cerebral tissue removal: algorithm and analysis. NeuroImage. 2010;56(4):1982–1992. doi: 10.1016/j.neuroimage.2011.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coupe P, et al. Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. NeuroImage. 2011;54:940–954. doi: 10.1016/j.neuroimage.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 6.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- 7.Ellingsen LM, et al. Segmentation and labeling of the ventricular system in normal pressure hydrocephalus using patch-based tissue classification and multi-atlas labeling. Proceedings of SPIE Medical Imaging (SPIE-MI 2016); San Diego, CA. 27 February–3 March 2016; 2016. pp. 97840G–97840G-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischl B. FreeSurfer. NeuroImage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ledig C, et al. Robust whole-brain segmentation: application to traumatic brain injury. Med Image Anal. 2015;21:40–58. doi: 10.1016/j.media.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Poca MA, et al. Is the placement of shunts in patients with idiopathic normal pressure hydrocephalus worth the risk? Results of a study based on continuous monitoring of intracranial pressure. J Neurosurg. 2004;100(5):855–866. doi: 10.3171/jns.2004.100.5.0855. [DOI] [PubMed] [Google Scholar]
- 11.Roy S, Carass A, Prince JL, Pham DL. Subject specific sparse dictionary learning for atlas based brain MRI segmentation. In: Wu G, Zhang D, Zhou L, editors. MLMI 2014 LNCS. Vol. 8679. Springer, Cham; 2014. pp. 248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roy S, et al. Subject-specific sparse dictionary learning for atlas-based brain MRI segmentation. IEEE J Biomed Health Inform. 2015;19(5):1598–1609. doi: 10.1109/JBHI.2015.2439242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiee N, Bazin PL, Cuzzocreo JL, Blitz A, Pham DL. Segmentation of brain images using adaptive atlases with application to ventriculomegaly. In: Székely G, Hahn HK, editors. IPMI 2011 LNCS. Vol. 6801. Springer; Heidelberg: 2011. pp. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toma AK, et al. Systematic review of the outcome of shunt surgery in idiopathic normal-pressure hydrocephalus. Acta Neurochir. 2013;155:1977–1980. doi: 10.1007/s00701-013-1835-5. [DOI] [PubMed] [Google Scholar]
- 15.Van Leemput K, et al. Automated model-based tissue classification of MR images of the brain. IEEE Trans Med Imaging. 1999;18:897–908. doi: 10.1109/42.811270. [DOI] [PubMed] [Google Scholar]
- 16.Wilcoxon F. Individual comparisons by ranking methods. Biom Bull. 1945;1(6):80–83. [Google Scholar]
- 17.Zhang J. The mean field theory in EM procedures for Markov random fields. IEEE Trans Signal Process. 1992;40:2570–2583. doi: 10.1109/83.210863. [DOI] [PubMed] [Google Scholar]


