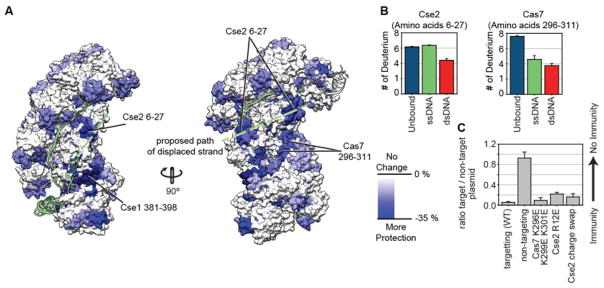
Figure 3. Using HDX to identify residues involved in binding the displaced DNA strand.
(A) Differences in deuterium uptake between the unbound and dsDNA bound complex for individual peptides at the 180-minute time point are mapped onto the dsDNA bound structure . Only peptides that are more protected in the dsDNA bound complex (blue) are shown. The proposed path of the displaced strand is indicated (green). (B) Deuterium incorporation for peptides Cse2 6–27 and Cas7 296–311 at the 180-minute time point is displayed for Cascade without DNA (teal), ssDNA bound (green), and dsDNA bound complexes (red). The Cse2 peptide shows increased protection in the dsDNA bound complex, as compared to the unbound or the ssDNA bound complex (p<0.01). The Cas7 peptide shows the most protection in the dsDNA bound complex. Error bars represent the standard deviation of three replicates. (C) HDX-guided mutation of positively charged residues on Cse2 (R12E), Cse2 (R53D, K77D, K78D, R89D, R135D, R143D and K158D, named Cse2 charge swap), and Cas7 (K296E, K299E, K301E) were tested in a plasmid-curing assay to determine the biological impact of mutating these residues. The Cas7 mutant did not have a significant impact while the Cse2 mutants displayed a small, but significant defect. The single mutation of Cse2 R12E had an equal impact as mutation of seven Cse2 residues. The error bars represent the standard error of the mean of three replicates.