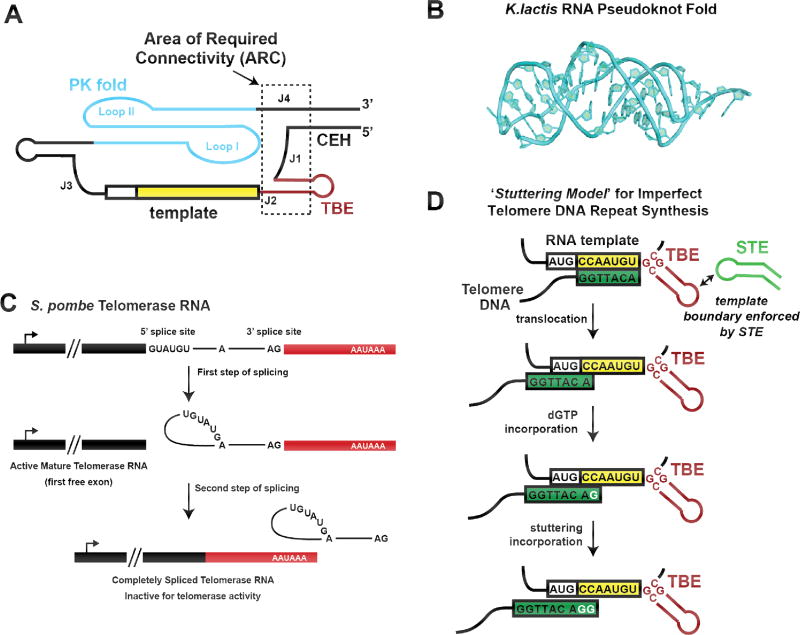
Figure 3. RNA structure and function in yeast telomerase.
(A) Schematic model of the core domain of yeast telomerase RNA, highlighting the area of required connectivity (ARC) outlined in dashed black box. Disruption of the RNA backbone in this region of the telomerase RNA disrupts catalytic activity. (B) High-resolution structure of the RNA pseudoknot fold from the budding yeast Kluyveromyces lactis. Figure is adapted from (56) PDB 2M8K. (C) Model for partial splicing during maturation of fission yeast telomerase RNA. After recognition by the yeast spliceosome, the first step of splicing produces a functional telomerase RNA with a mature 3’ end. The second step of splicing is highly inefficient and usually abortive for this transcript, but when it does proceed, the splicing product is an inactive telomerase RNA that is quickly degraded. (D) Model for active site ‘stuttering’ during yeast telomere DNA repeat synthesis. Many species of yeast possess irregular telomere repeat sequences. In the ‘Stuttering Model’, efficient template boundary definition is reinforced by the distal stem terminus element (STE), preventing run on reverse transcription beyond the template. After realignment of the 3’ end of the DNA substrate, the RT active site can incorporate extra dGTP nucleotides, resulting in telomere repeats of varying length and sequence.