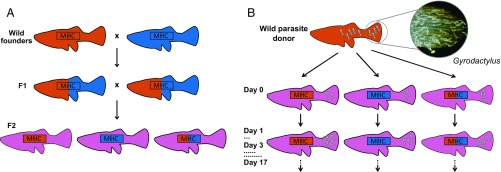
Fig. 1.
Schematic of the experiment. (A) Breeding design. Wild fish from two populations (P-generation) were crossed to produce F1s that were heterozygous across the genome with respect to population of origin. These were allowed to mate at random to produce F2s that segregated into heterozygotes and two types of homozygotes at the focal MHC class II locus, while having, on average, “mixed” genetic backgrounds [23 chromosome pairs (40), plus crossing-over when F1s reproduce]. (B) Controlled experimental infections. Two gyrodactylid worms from one of the P-generation source streams were inoculated on to the caudal fin of each F2 fish. Each infected fish was then kept in isolation and its infection monitored every other day for 17 d (22).