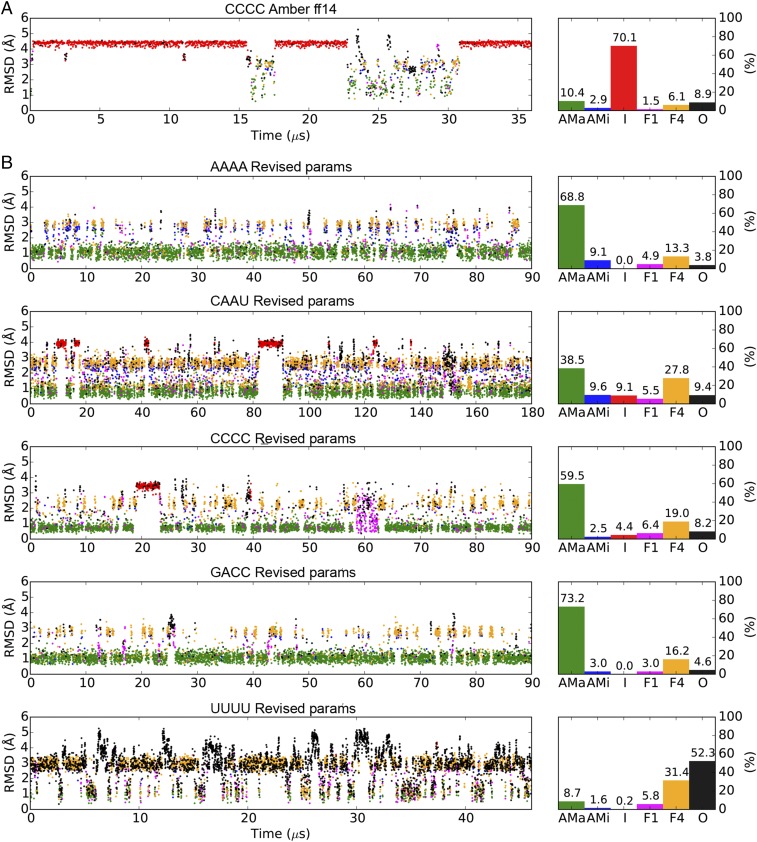
Fig. 2.
MD simulations of ssRNA tetramers using AMBER ff14 and the TIP3P water model (A) or the revised parameters (B). The scatter plots on the left indicate rmsds of the backbone (including the ribose) nonhydrogen atoms to the initial A-form structures. Based on the stacking patterns of the four nucleotides, the simulated conformations are categorized into six groups: A-form major (AMa, green), A-form minor (AMi, blue), Intercalated (I, red) Nucleotide-4 flipped (F4, yellow), Nucleotide-1 flipped (F1, magenta), and Others (O, black). See Methods for the detailed definitions of these conformations. Percentage populations of the six conformations in each simulation are shown on the right.