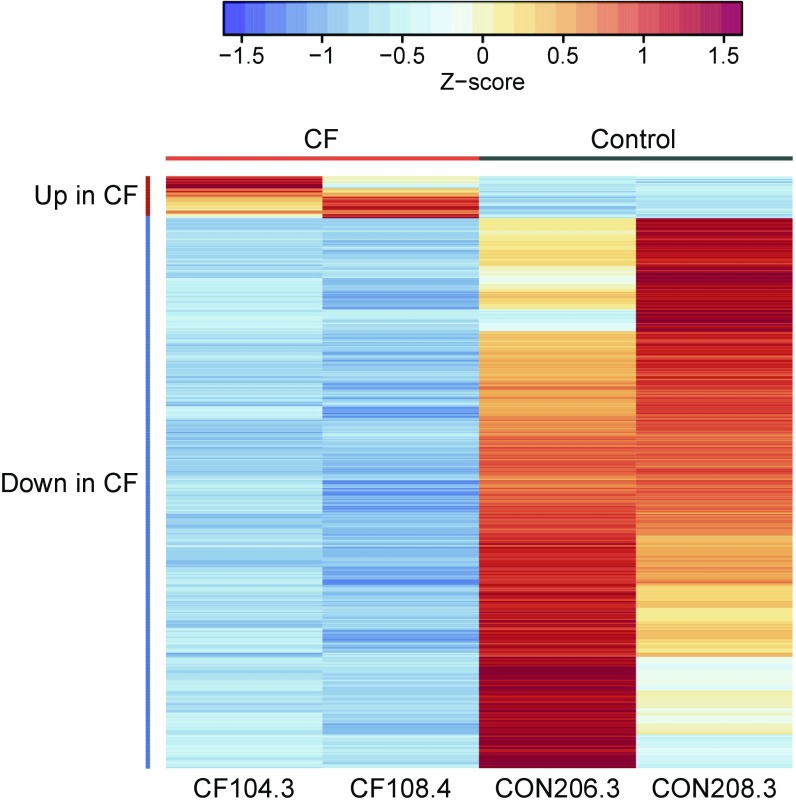
Fig. 3.
DEGs in GlyMM in CF E. coli isolates relative to controls are predominantly down-regulated. Normalized absolute expression z-scores for each gene and isolate were calculated from gene-wise variance estimates using DESeq2. Pearson correlation was used to cluster genes (rows). Genes up in CF include translation factors, TCA, amino acid synthesis, and energy production (tufB, tufA, atpCD, rspEG, rpmD, aceE, metE, sucBC, hisA, and ilvE). Genes down in CF are involved in biofilm, stress, and acid resistance (azuC, ymgC, cadB, gadBCEX, hdeABD, sra, arfA, ecpCDR, rcsA, csgBDG, ariR, psiE, ymgA, cspGH, bdm, adiCY, ycgZ, rclAC, ynhA, yjbJ, yhcLN, and ygiW). Only genes that were differentially expressed between the CF and control (CON) E. coli isolates are shown.