Cell division is an essential step in the life of all cells. Fungi, amoeboid, and mammalian cells divide by the assembly and constriction of a contractile ring of actin, myosin, and other highly conserved proteins (1). The mechanism of cytokinesis is best understood in the fission yeast Schizosaccharomyces pombe. Combined years of research have identified more than 100 genes involved in building and constricting the contractile ring (1). Quantitative fluorescence imaging techniques have been developed to calculate the local and global concentration of endogenously tagged proteins involved in cytokinesis (2, 3). Mathematical models were built using these quantitative data to explain the principles that govern the mechanisms of contractile ring assembly (4) and constriction (5). These mathematical models were limited by the lack of information about the organization of proteins within the contractile ring.
How the proteins involved in building and constricting the contractile ring organize to generate tension force is still vastly unknown. Recently, superresolution imaging has enabled us to capture the molecular organization of proteins within the constricting contractile ring in fixed cells (6), and high-speed superresolution imaging combined with quantitative analyses has provided molecular organization of protein complexes within the assembling and constricting contractile ring in live cells (7). Even with this combined knowledge, there are still many aspects of the general mechanisms of contractile ring assembly, constriction, and disassembly that we do not understand (8). One remaining enigma about cytokinesis is the how the network of actin filaments, the most abundant protein of the contractile ring, is structured during constriction. Actin filaments within the contractile ring are polymerized, severed, depolymerized, and pulled upon by the action of many other actin-binding proteins present in the cytokinetic machinery. Together, these modifications to the actin network generate tension force that is transmitted to the ingressing plasma membrane via a poorly understood and likely complex anchoring mechanism. Visualizing the precise organization of single-actin filaments within the contractile ring has the potential to answer questions about the abundance of actin within the ring, how the actin filaments interact with one another, and how they connect to the plasma membrane to bring us one step closer to understanding the general mechanism of cytokinesis. However, visualizing single-actin filaments within the dense actin bundle is deceptively complicated. The bundle is too dense for actin filaments to be resolved by traditional confocal microscopy or even by superresolution microscopy. Electron microscopy efforts have generated too few datasets to provide informative insight into the organization of actin filaments within the contractile ring (9, 10).
In PNAS, Swulius et al. (11) used electron cryotomography of cryopreserved dividing fission yeast cells to generate exquisite 3D reconstruction images of thick sections of the constricting contractile ring (Fig. 1). Using cdc25-22, a commonly used cell cycle mutation, the authors generated synchronized populations of dividing fission yeast cells exhibiting constricting contractile rings. After cryopreservation, Swulius et al. used cryosectioning or cryofocused ion beam milling to produce ∼200-nm-thick sections through the contractile ring. Tilt series of the sections were imaged by electron cryotomography and the resulting datasets were reconstructed into stunning 3D images of sections of the contractile ring containing the ingressing plasma membrane and single-actin filaments inside the contractile ring bundle. These 3D reconstructions of transverse sections through contractile rings contain a wealth of information for quantitative analyses. Arrays of ∼7.5-nm-wide actin filaments can be identified over the dense cytoplasmic background at the leading front and also sometimes slightly off and up against the side edges of the ingressing plasma membrane. The filaments continuously traverse the 200-nm-thick sections, confirming their identity as actin filaments over cytoplasmic protein background noise.
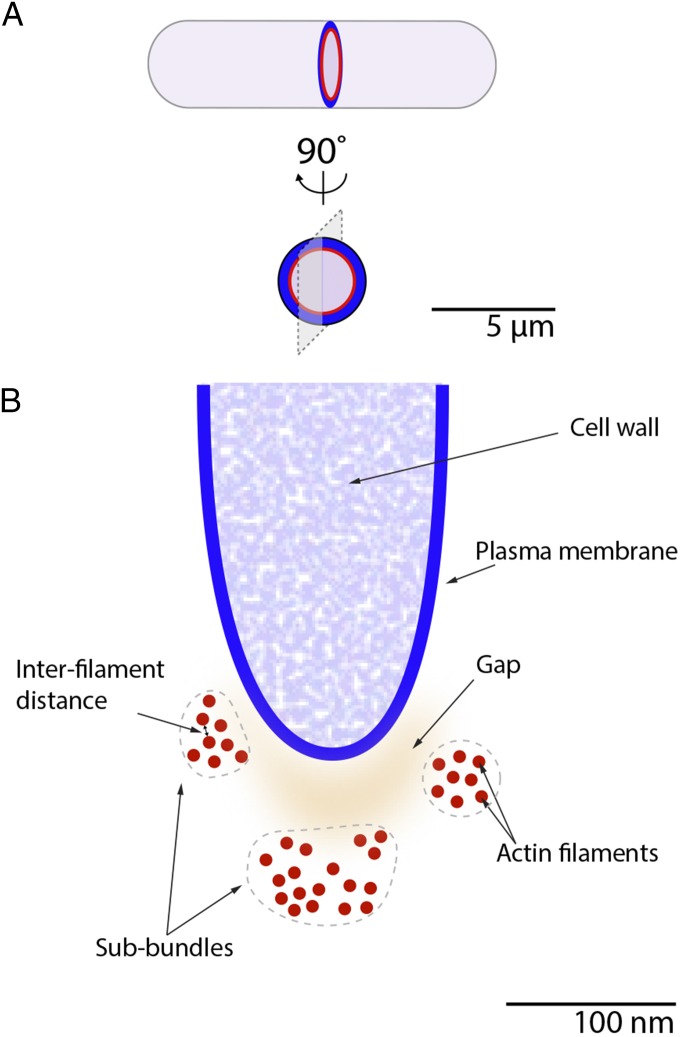
Fig. 1.
Conceptual model summarizing the main features of the organization of the actin filaments within the contractile ring. (A) Schematic diagram illustrating the orientation of the 3D reconstruction. Diagram of the longitudinal view (Upper) and face view (Lower) of a fission yeast cell with a ring of actin filaments (red) at the front edge of the ingressing plasma membrane (blue). Gray plane illustrates the orientation of the transverse section through the ring shown in B. (B) Schematic diagram illustrating the transverse section of a contractile ring with the plasma membrane (blue) and three subbundles (surrounded by gray outlines) of actin filaments (red). Features are drawn to scale. The separation between filaments (interfilament distance) is variable with a peak value at 4.5 nm. The gap between the plasma membrane and the edge of the actin filament varies in size (orange shading).
Visual comparison of the reconstructions illustrated in the work of Swulius et al. (11) is sufficient to notice the high level of variability in the overall appearance of the bundle of actin filament between the samples. The number of filaments per sample, the packing of the actin filaments within the bundle, and the position of the bundle with respect to the plasma membrane vary greatly from one reconstruction to the next. Such diversity in the organization of the actin filaments seemingly rules out that the filaments of the contractile ring are organized in a highly regular lattice, such as the highly ordered organization of actin filaments of the muscle sarcomere. The number of actin filaments per section ranges from 14 to 60, distributed in a bundle of ∼150 nm in diameter, consistent with previously published measurements (7, 12). Assuming a uniform distribution of the actin filaments around the ring, Swulius et al. (11) calculated that the average length of the actin filaments is nearly 1 µm in length with ∼285 filaments per ring. Measurements of the distance separating actin filaments from their nearest neighbor result in a distribution of values with a peak value corresponding to a distance of 4.5 nm from the edge of one filament to its nearest neighbor. This narrow spacing between actin filaments leaves little space for cross-linking proteins, only about the size of the one actin-binding domain of α-actinin. Larger actin filament binding proteins may therefore preferentially localize to larger spaces between filaments within the bundle. In some reconstructions, the bundle was separated into subbundles of actin filaments by large ∼22-nm gaps. The reason for the presence of these gaps is unknown but the magnitude of their size is too great for most actin cross-linking proteins to link these subbundles together. The large gaps may be the outcome of the dynamic events that occur within the bundle as a result of constriction or turnover of the actin filaments.
Filaments within the reconstructed sections run rather parallel to one another and measurements show that they are quite straight over the ∼200-nm sections. Although this technique cannot distinguish the pointed from the barbed ends of the actin filaments, filament ends are detected throughout the entire thickness of the sections. Through careful observation of the reconstructions, it was found that the ends of filaments are distributed randomly across the thickness of the bundle and thus do not tend to gather into groups that may have indicated filament polymerization or anchoring “hubs.”
The shape of the bundle of actin filaments follows the shape of the plasma membrane, or “saddles” the ingressing plasma membrane, yet direct contact between the bundle and the plasma membrane was not observed in any of the reconstructions. From observing the reconstructions, the size of the gap between the plasma membrane and closest edge of the bundle appears quite variable, ranging from ∼10–60 nm. These observations suggest the presence of an anchoring mechanism within that gap that links the constricting actin filament bundle to the plasma membrane. This anchoring mechanism may function to relay the contractile force generated within the ring to the plasma membrane. The protein complexes involved in this mechanism would need to be flexible either in size or in the molecular conformations they can adopt to accommodate the varying distance between the plasma membrane and the bundle of actin filaments. Unfortunately, protein anchors linking the plasma membrane to the actin bundle were not distinguishable in these datasets.
No other proteins known to be present in the contractile ring could be identified or resolved in the datasets obtained from this technique. Myosin type-II molecules were not observed between the actin filaments and no obvious protein structure could be distinguished linking the actin bundle to the plasma membrane. It is likely that these other proteins are preserved during the cryopreservation step but without any particularly ordered structural features these other proteins cannot be distinguished during the analysis steps from cytoplasmic proteins present in the images. Surprisingly, however, other actin structures were not visualized in these 3D reconstructions. Thin, dynamic actin bundles flanking the main central bundle of the contractile ring that are visible in live cells imaged by confocal fluorescence microscopy and high-speed superresolution microscopy (5, 7) were not captured in these reconstructions. These thin bundles are well separated and often run at sharp angles from the main central bundle of actin, which could explain why they were not captured during the analysis.
Swulius et al. (11) see no correlation either between the calculated filament length and the ring diameter or between the calculated number of filaments in the ring and the ring diameter. These measurements would suggest that the number of filaments and their length do not vary during the constriction of the contractile ring. If this were true, actin would accumulate as the ring constricts and the actin density in the ring would rise with decreasing ring diameter. This is contradictory to fluorescence measurements (12), which showed that the amount of actin decreases as the ring constricts and actin density is maintained throughout constriction. The high variability in the number of actin filaments of the bundle combined with the small sample size for rings at each step of constriction could explain this discrepancy.
In combination with quantitative fluorescence imaging and high-speed superresolution techniques, electron cryotomography of cryopreserved samples adds a powerful tool to our visualization toolkit in the quest to elucidate the mechanisms that govern cytokinesis. This method could be used to image cells mutant for different genes involved in cytokinesis and help identify how they affect the bundle of actin filament during contractile ring constriction. For example, cells mutant for myo2, one of the two type II myosins involved in cytokinesis, exhibit a broader, less compact actin bundle, but it is not known whether this effect is due to a looser packing of the actin filaments, the creation of more large gaps between subbundles, or even perhaps the presence of more actin filaments in the bundle (13). The process of ring disassembly is another poorly understood step of cytokinesis. Electron cryotomography of cryopreserved samples could be used to visualize the actin network of disassembling contractile ring in cells that are nearly completing ring constriction, which would likely provide valuable information about the mechanisms that break down and clear the contractile ring before cell separation.
Footnotes
The author declares no conflict of interest.
See companion article on page E1455.
References
- 1.Pollard TD, Wu JQ. Understanding cytokinesis: Lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 3.Wu JQ, McCormick CD, Pollard TD. Chapter 9: Counting proteins in living cells by quantitative fluorescence microscopy with internal standards. Methods Cell Biol. 2008;89:253–273. doi: 10.1016/S0091-679X(08)00609-2. [DOI] [PubMed] [Google Scholar]
- 4.Vavylonis D, Wu JQ, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 5.Stachowiak MR, et al. Mechanism of cytokinetic contractile ring constriction in fission yeast. Dev Cell. 2014;29:547–561. doi: 10.1016/j.devcel.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDonald NA, Lind AL, Smith SE, Li R, Gould KL. Nanoscale architecture of the Schizosaccharomyces pombe contractile ring. eLife. 2017;6:6. doi: 10.7554/eLife.28865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laplante C, Huang F, Tebbs IR, Bewersdorf J, Pollard TD. Molecular organization of cytokinesis nodes and contractile rings by super-resolution fluorescence microscopy of live fission yeast. Proc Natl Acad Sci USA. 2016;113:E5876–E5885. doi: 10.1073/pnas.1608252113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollard TD. Nine unanswered questions about cytokinesis. J Cell Biol. 2017;216:3007–3016. doi: 10.1083/jcb.201612068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamasaki T, Osumi M, Mabuchi I. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J Cell Biol. 2007;178:765–771. doi: 10.1083/jcb.200612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanbe T, Kobayashi I, Tanaka K. Dynamics of cytoplasmic organelles in the cell cycle of the fission yeast Schizosaccharomyces pombe: Three-dimensional reconstruction from serial sections. J Cell Sci. 1989;94:647–656. doi: 10.1242/jcs.94.4.647. [DOI] [PubMed] [Google Scholar]
- 11.Swulius MT, et al. Structure of the fission yeast actomyosin ring during constriction. Proc Natl Acad Sci USA. 2018;115:E1455–E1464. doi: 10.1073/pnas.1711218115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courtemanche N, Pollard TD, Chen Q. Avoiding artefacts when counting polymerized actin in live cells with LifeAct fused to fluorescent proteins. Nat Cell Biol. 2016;18:676–683. doi: 10.1038/ncb3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laplante C, et al. Three myosins contribute uniquely to the assembly and constriction of the fission yeast cytokinetic contractile ring. Curr Biol. 2015;25:1955–1965. doi: 10.1016/j.cub.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]