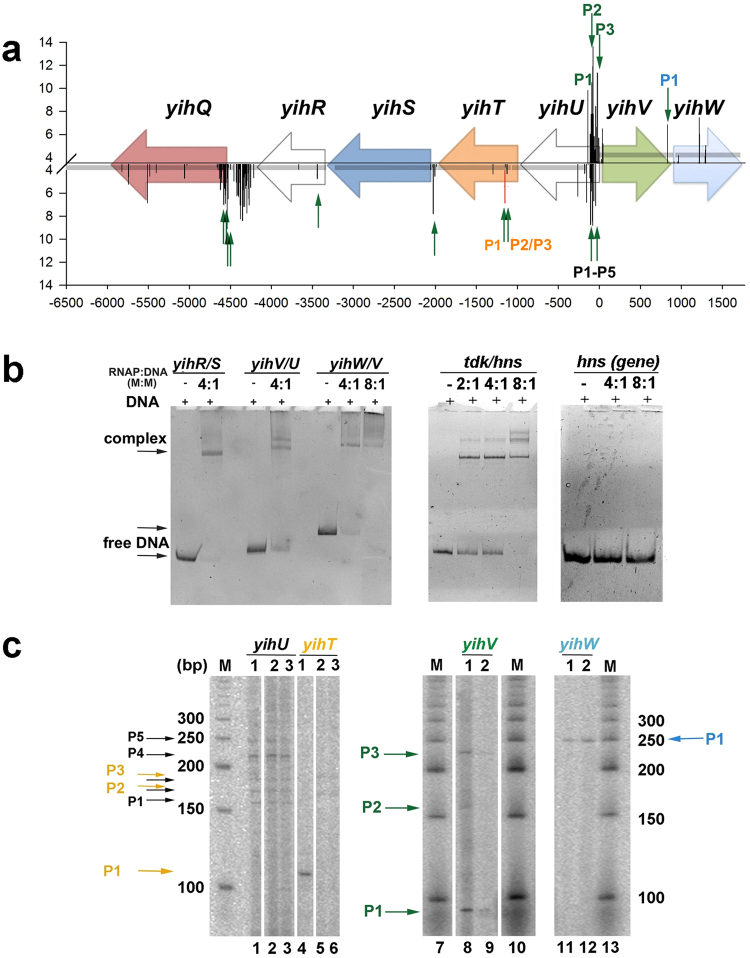
Figure 3.
Promoter mapping within the yih cassette. (a) Schematic representation of the yih genes and candidate promoters predicted in silico. Horizontal arrows show gene positioning. Bars represent the transcription start points predicted by PlatProm on both DNA strands, their scores are indicated on the Y axis. Score assigned to the yihTP1 promoter by the unified algorithm, PlatPromU, is marked with red. X axis represents positioning related to the yihV ATG start codon. Green vertical arrows point out the 5′-end of RNA detected by the 5′-end-specific RNA-Seq31. (b) Band-shift assays with σ70-RNA polymerase suggests the presence of single promoters in the intergenic regions yihV/W and yihS/R, and two promoters in yihU/V. Molar ratios are indicated above the lanes. All samples were run on one gel. Known σ70-promoter of the hns gene was used as a positive control; intergenic region within hns where no promoters were predicted was used as a negative control (c) Primer extension analysis revealed transcription start point for yihW and several starts for the yihU, yihT, and yihV genes, some of which were activated during growth on lactose. (1) Growth on lactose, (2) growth on glucose, (3) growth on glycerol. The yihU and yihT samples, as well as the DNA ladder on the left, were run on one gel, while yihV and yihW were run on another gel, together with the DNA ladders displayed. Lanes that were taken from different parts of the gels are divided by spacing.