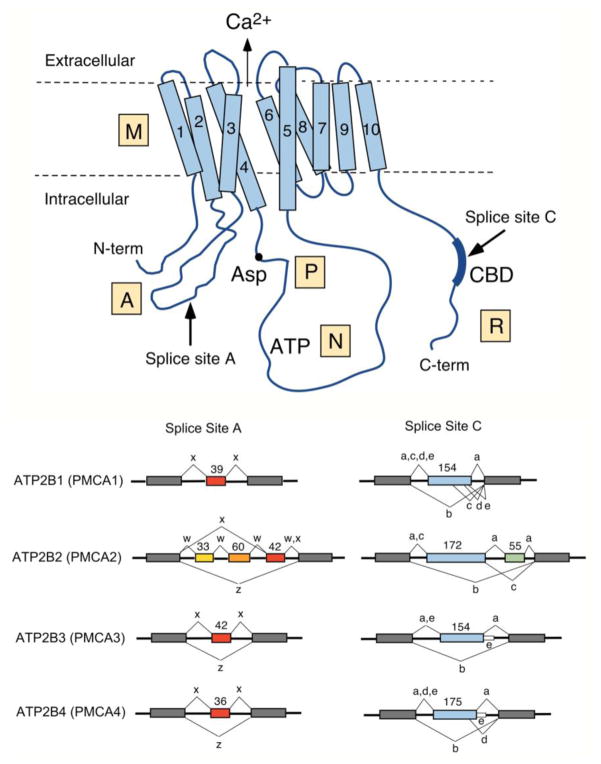
Figure 1. Scheme of the PMCA and major alternative splice pattern of human ATP2B genes.
Top: Schematic representation of the PMCA with major domains indicated M, membrane; A, actuator, P, phosphorylation; N, nucleotide-binding; and R, regulatory domain. The ten membrane-spanning regions are numbered, the N- and C-terminal ends are indicated, and the conserved aspartate (Asp) residue phosphorylated during the reaction cycle, the catalytic ATP-binding site (ATP), and the CaM-binding region (CBD) are labeled. The direction of Ca2+ transport is indicated by an arrow. Arrows also mark the positions where alternative splicing leads to isoform diversity at sites A and C. Bottom: Splicing options of human PMCA genes ATP2B1-ATP2B4. The exon structure in the region of alternative splicing at sites A and C is shown for each ATB2B (PMCA) gene. Constitutively spliced exons are represented by gray boxes, alternatively spliced exons are colored and their size is shown in nucleotides. Splice options are indicated by connecting lines, and the resulting splice variants are labeled by their lowercase symbol. Note that only splice option “x” has been found in PMCA1 at site A, and that variant “e” in PMCA3 and PMCA4 results from a read-through of the last alternatively spliced exon into the following intron (shown as thin white box).