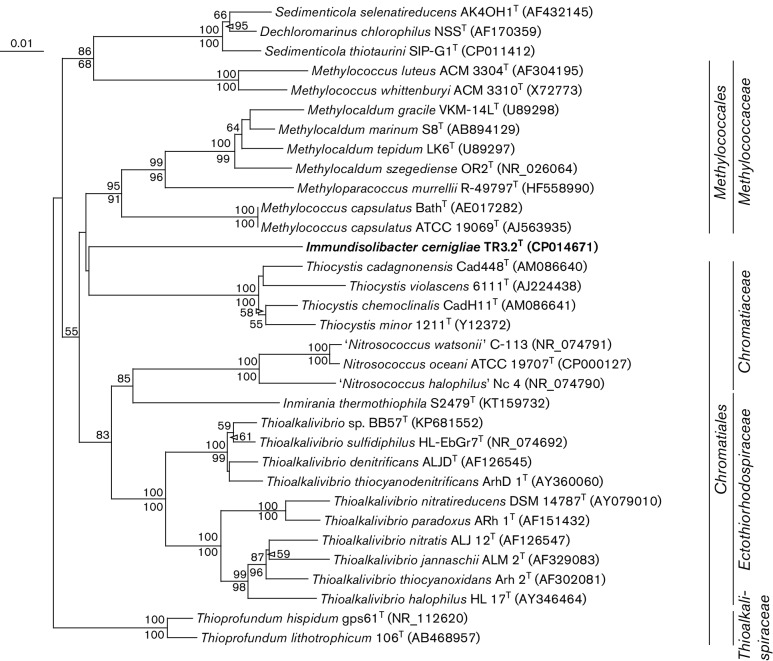
Fig. 2.
16S rRNA gene neighbour-joining phylogenetic tree of strain TR3.2T with 16S rRNA gene sequences of the most similar cultivated and described strains. Sequences were aligned and the neighbour-joining tree created using clustalx 2.1 [46] and without considering positions with gaps. The maximum-parsimony tree was determined using the paup [47] implementation within Geneious 9.1.3 [48] and the same alignment file generated for the neighbour-joining tree. Bootstrap values for neighbour-joining and maximum-parsimony algorithms are presented as a percentage of 1000 iterations above and below nodes, respectively. GenBank accession numbers are presented in parentheses. The order and family of cultivated strains are indicated on the right side. Pseudomonas putida strain IAM 1236 (accession no. D84020) was used as an outgroup (not shown). Bar, 0.01 substitutions per nucleotide position.