Fig. 1.
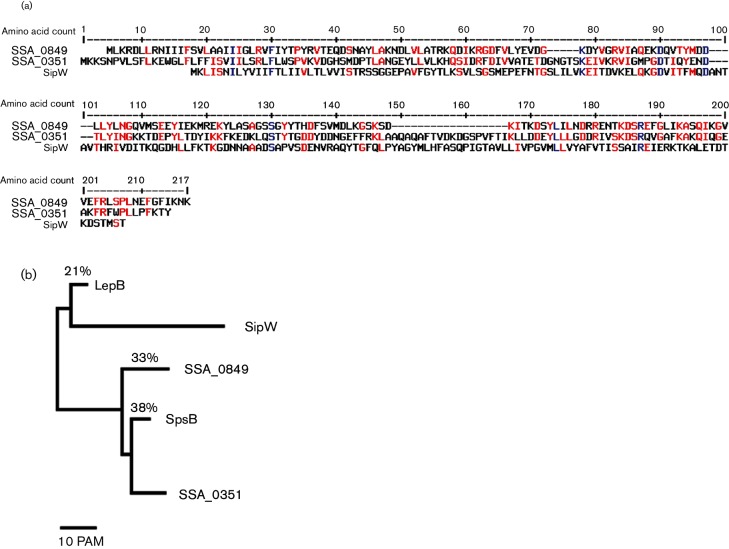
Characteristics of SSA_0351-encoded SPase as extracted from various databases. (a) Multiple protein sequence alignments of S. sanguinis SPases, SSA_0351 and SSA_0849, and B. subtilis SipW were conducted using MultAlin. Blue: identity across three sequences. Red: identity between two sequences. (b) Phylogenic tree showing the evolutionary relatedness of bacterial SPases (LepB from Escherichia coli str. K-12 substr. MG1655, SpsB from Staphylococcus aureus subsp. aureus NCTC 8325, SipW from Bacillus subtilis) based on protein sequence alignment with hierarchical clustering. The identity percentages of the protein sequence alignments for every SPase with respect to SSA_0351 are shown, except for SpiW, which was too low. The evolutionary distances between SPases have been calibrated to 10 PAM, where 1 per cent accepted mutation (PAM) is the time in which 1 substitution event per 100 sites is expected to have happened.