Figure 1.
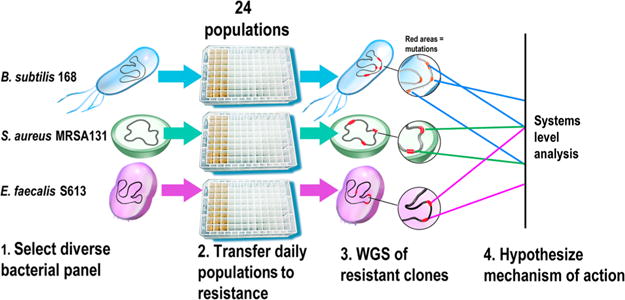
Summary of approach to identify the mechanism of action of a novel antibiotic. Identifying an antibiotic’s mechanism of action can be challenging, as antibiotics can target a diverse range of processes in bacteria, such as cell wall synthesis, protein translation, and nucleic acid metabolism. Therefore, we developed an approach that can be used to efficiently develop a hypothesis for the mechanism of action of any novel antibiotic. (1) A panel of at least three different susceptible species of bacteria is selected on the basis of several different criteria. Ideally, the different strains should have well-annotated genomes that would allow mutated genes and pathways to be readily identified. Model organisms also allow experimental validation of the proposed mechanism. Thus, it is beneficial to include at least one model organism, such as a laboratory strain of Bacillus subtilis or Escherichia coli. Additionally, the panel should consist of phylogenetically diverse species, since distantly related organisms are more likely to adapt through different mechanisms and thus reveal more pathways and targets affected by the antibiotic. Finally, some of the selected strains should be clinically relevant species that the antibiotic would potentially be used against. (2) Replicate populations of each species are adapted to become resistant to the antibiotic under study. Including replicate populations of the same strain is important since it allows for the identification of genes and pathways that are mutated reproducibility across the different populations, which in turn reveals which targets are most important to resistance. During adaptation, conservative increases in antibiotic concentration should be used to favor genetically diverse populations. (3) Comparative WGS between the adapted resistant strains and the ancestral susceptible starting strains is used to determine what adaptive mutations were acquired. (4) The different adaptive mutations are compared across replicate populations and the different species to develop a hypothesis for the mechanism of action of the antibiotic. Loci and pathways mutated across the different replicate populations and species are more likely to be important to resistance and thus targets of the antibiotic. Experimental validation is then performed to confirm the proposed mechanism of action.
