Fig. 3. DnaJ binding to the unfolded ubiquitin is force-dependent.
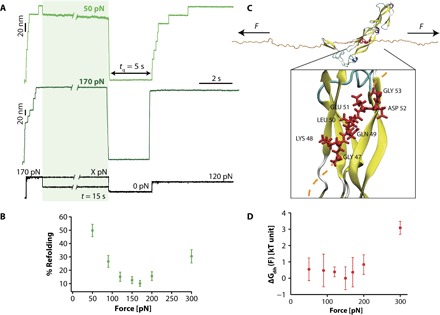
(A) Changing the force at which ubiquitin is left stretched after mechanical unfolding results in a marked change in the refolding yield. In these experiments, a high force of 170 pN for a short time t = 1 s triggers ubiquitin unfolding. The force is then varied within the range X = 50 to 300 pN for a period t = 15 s before a tq = 5 s pulse is applied to trigger protein refolding. The refolding efficiency is tested in the test pulse, when the protein is stretched back at 120 pN. (B) Dependency of the refolding percentage with the pulling force (F = 30 pN, n = 33; F = 50 pN, n = 30; F = 90 pN, n = 25; F = 120 pN, n = 36; F = 150 pN, n = 28; F = 170 pN, n = 45; F = 200 pN, n = 43; F = 300 pN, n = 22). (C) Snapshot of the interaction of DnaJ (PDB: 1NLT) with the unfolded and mechanically stretched ubiquitin (orange ribbon): The interaction surface involves seven residues (red), exhibiting the consensus sequence GKQLEDG. Inset: Zoom of the reconstructed (see Materials and Methods) DnaJ-ubiquitin fragment system. The bonds of the amino acids from the consensus sequence are shown as red tubes, and the corresponding Cα’s are shown as red balls. (D) Five couples (ϕ, ψ) of backbone dihedral angles can be defined for the consensus sequence to estimate the free-energy contribution corresponding to the overall energetic cost underlying the force-induced remodeling of the dihedral angles of the ubiquitin interaction fragment upon DnaJ binding (see Materials and Methods and the Supplementary Materials for details about the calculation). Error bars correspond to the SDs observed among the five independent runs at each force, and the data are normalized with respect to the lowest free-energy value (observed at 150 pN).
