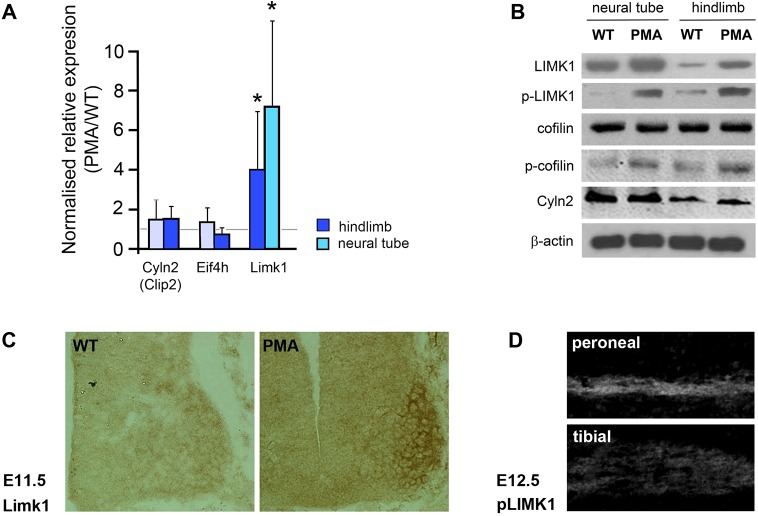
Fig. 8.
Upregulation of Lim-kinase 1 (Limk1) in pma/pma mice. (A) qPCR data of the top-three candidate ‘clubfoot’ genes in the PMA mouse: Cyln2, Eif4h and Limk1. All data are means from three independent replicates, comparing E11.5 pma/pma homozygotes with C57BL/6 controls. cDNA was prepared separately from hindlimbs and neural tube of both genotypes. The error bars represent the 95% confidence interval from the statistical randomisation method from the REST software. Limk1 transcripts were significantly over-represented in both hindlimbs and the neural tube of pma/pma homozygotes. *P<0.05. (B) Western blot of E11.5 neural tube and hindlimbs of wild-type (WT) and pma/pma embryos. Limk1 and p-Limk1 were present at higher levels in pma/pma homozygotes. Total levels of cofilin (the main substrate of Limk1) were not affected but levels of phosphorylated cofilin increased, as would be expected with upregulation of Limk1. Cyln2 was not affected by pma genotype. β-actin was used as the loading control. (C) Immunohistochemistry for Limk1 (brown) in the E11.5 neural tube of wild-type and pma/pma embryos. Limk1 is present at higher levels in PMA mice, especially in the medial and lateral LMC. (D) Immunohistochemistry for p-Limk1 in dorsal (peroneal) and ventral (tibial) branches of the sciatic nerve at E12.5. p-Limk1 is present at higher levels in the developing peroneal nerve. See also Fig. S11.