Fig. 2.
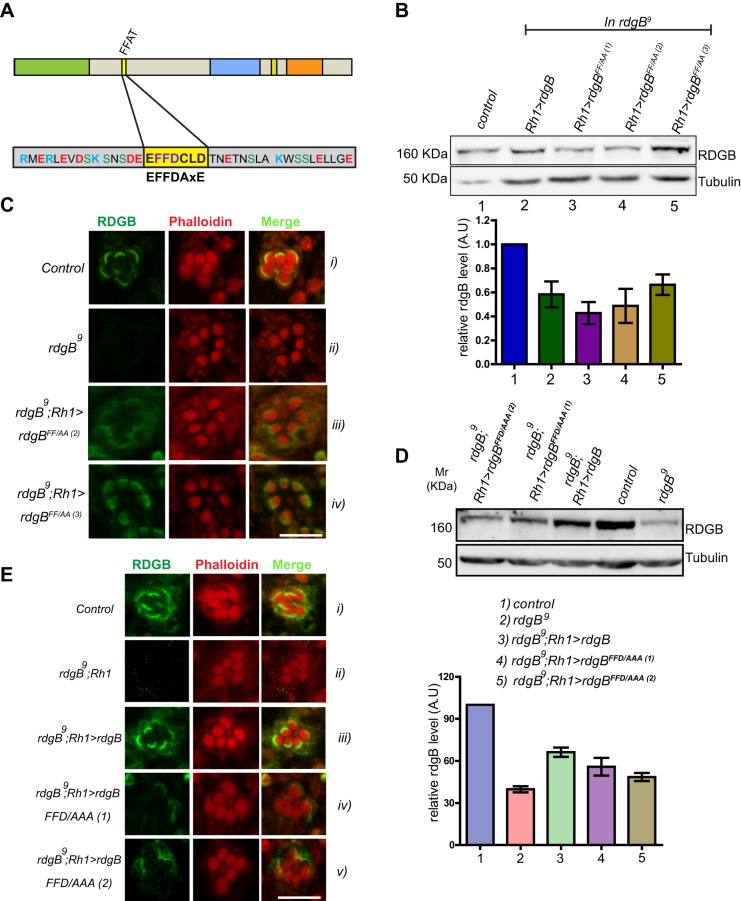
An intact FFAT motif is required to localize RDGBα to the SMC. FFAT mutants were expressed in rdgB9 flies using Rh1-Gal4 and their ability to rescue rdgB9 was tested. (A) Diagrammatic representation of various domains in RDGB (drawn to scale); PITPd, DDHD and LNS2 are three domains whereas DGBL refers to a DAG binding-like region. The core residues of the FFAT motif are highlighted in yellow, along with flanking residues. The FFAT motif was mutated to disable the ability of the FFAT motif to bind to VAP. Two versions were tested: FF→AA (RDGBFF/AA) and FFD→AAA (RDGBFFD/AAA). (B) Representative immunoblot showing expression level of RDGB in different RDGBFF/AA transgenic lines (top panel). Blot was re-probed for tubulin as a loading control. Bar chart shows the quantification of band intensity (n=2 blots). Error bars represent s.e.m. (C) Localization of the RDGBFF/AA protein in control, rdgB9 and two different transgenic lines. (D) Representative immunoblot showing expression level of RDGB in different RDGBFFD/AAA. Quantification of band intensity from these blots is shown in bar chart. Error bars represent s.e.m. (n=3 blots). (E) Confocal images showing localization of RDGBWT and RDGBFFD/AAA (two independent transgenic lines) in the background of rdgB9. Control and rdgB9 photoreceptors are also shown. Scale bars: 5 µm.