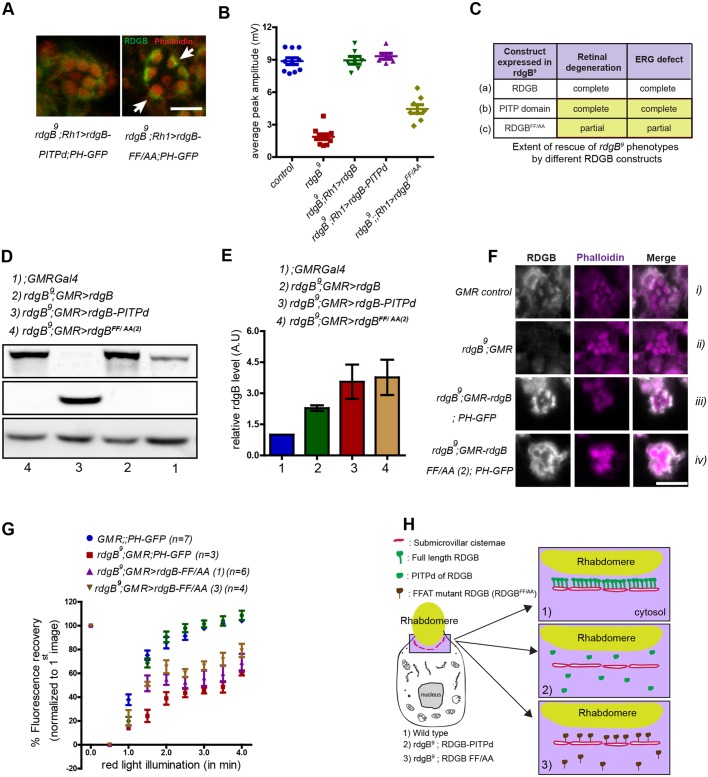
Fig. 7.
An intact FFAT motif is essential for RDGBα function independent of SMC localization. (A) High-magnification confocal images of a single ommatidium from rdgB9;Rh1>PITPd and rdgB9; Rh1>rdgBFF/AA. Note the relatively strong and concentrated staining near the base of rhabdomeres in the case of RDGBFF/AA compared with uniformly dispersed staining in case of PITPd. (B) Average ERG response of 1-day-old flies plotted from data pooled across experiments performed under the same conditions (for comparison). Error bars represent s.e.m. (C) Summary of rescue experiments using different RDGB constructs in rdgB9 flies. Compare the level of functional rescue achieved by expressing wild-type RDGB (a), only the PITPd of RDGB (b) and RDGB with mutated FFAT motif but intact wild-type PITP domain (c). Results are shown for equivalent level of protein expression. (D) Representative immunoblot image showing increase in expression level of RDGB and PITPd with GMR-Gal4 promoter. Tubulin was used as a loading control and rgdB9 is a negative control for RDGB. (E) Quantification of band intensity of RDGB normalized to tubulin as depicted in D (n=3 blots). Error bars represent s.e.m. (F) Confocal images showing localization of RDGBFF/AA in PRs using GMR-Gal4 promoter. Appropriate genetic controls were stained simultaneously. (G) Recovery kinetics of fluorescent pseudo pupil with time. Average fluorescence intensity of each pseudo pupil image is expressed as a percentage of the intensity of first image acquired. Each data point represents mean±s.e.m. (H) Cartoon representation (not to scale) of the localization and relative number of PITPd expressed in photoreceptors in case of wild-type flies (1), rdgB9 flies expressing PITPd (2) or RDGBFF/AA (3). Scale bars: 5 µm.