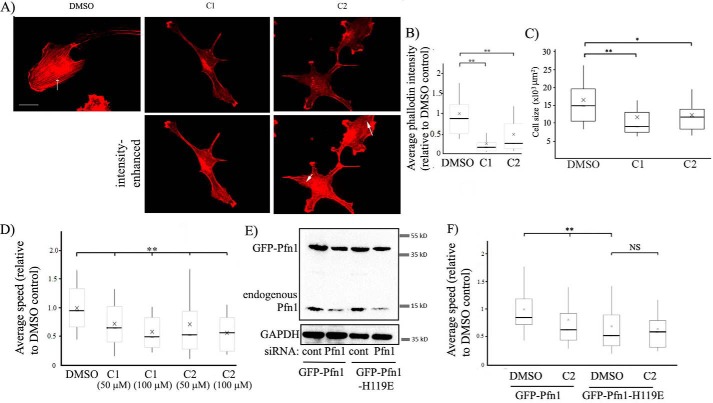
Figure 5.
Effects of a small-molecule inhibitor of Pfn1–actin interaction on the actin cytoskeleton and random 2D motility of ECs. A–C, representative fluorescence images (A) of rhodamine-phalloidin–stained HmVECs (acquired with a ×60 objective) following overnight treatment with either DMSO or C1 or C2 at 50 μm concentration (the arrow shows actin stress fibers; scale bar = 20 μm). The top panels show images at identical brightness/contrast settings. The images in the bottom panel were brightness-enhanced to reveal stress fibers (arrows) in compound-treated cells, albeit in much lower abundance than control cells. The box and whisker plot in B summarizes the mean values of the integrated fluorescence intensity of rhodamine–phalloidin relative to the average value of the DMSO control group. The mean values of the cell area for the indicated treatments are summarized in a box and whisker plot in C (n = 50–60 cells/group pooled from 3 experiments; **, p < 0.01; *, p < 0.05). In the box and whisker plots, X represent the mean, the middle lines of the box indicate the median, the top of the box indicates the 75th percentile, the bottom of the box measures the 25th percentile, and the two whiskers indicate the 10th and 90th percentiles, respectively. D, a box and whisker plot depicting the relative speed of randomly migrating HmVECs after treatment with DMSO or the indicated concentrations of compounds C1 or C2 (n = 100–120 cells/group pooled from 3 independent experiments; **, p < 0.01). E, Pfn1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, loading control (cont)) immunoblots of total extracts of HmVECs stably expressing either GFP–Pfn1 or GFP–Pfn1–H119E and following transfection with the indicated siRNAs. F, a box and whisker plot summarizing the effect of C2 on the relative speed of WT Pfn1 versus H119E Pfn1 expressers of HmVECs treated with control or Pfn1 siRNA (n = 60–70 cells/group pooled from 3 independent experiments; **, p < 0.01; NS, not significant).