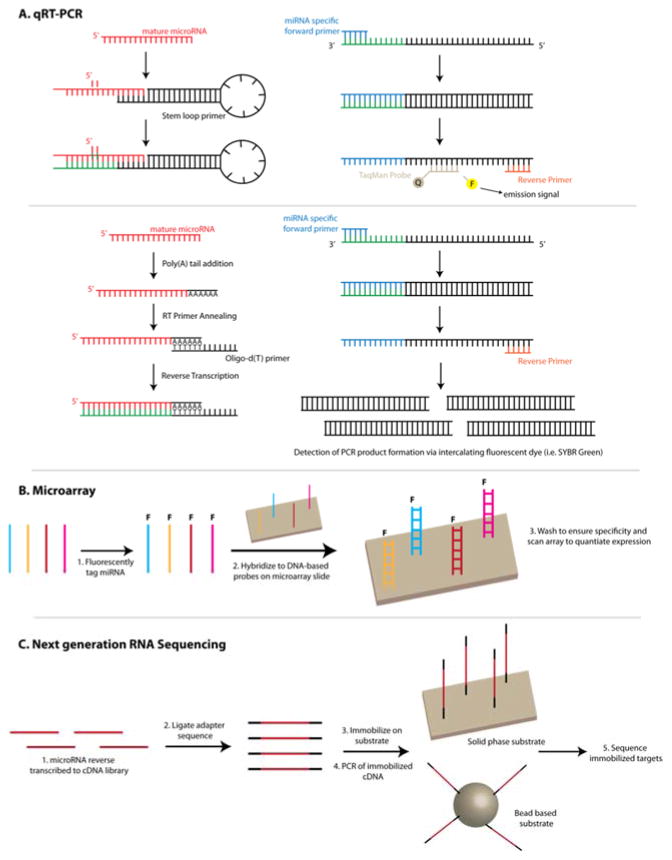
Figure 3.
Overview of conventional techniques: (a) qRT-PCR, (b) microarrays and (c) next generation RNA sequencing. (A) When using TaqMan qRT-PCR, the reverse transcription process utilizes stem-loop primers specific to the miRNA target of interest. During PCR amplification, the DNA polymerase proceeds along the template strands produces by miRNA specific forward and reverse primers and hydrolyses the TaqMan probe bound to the template. This liberates the fluorescent dye from the quencher and results in light emission. In SYBR green-based approaches, miRNAs are typically polyadenylated at the 3′ end and d(T) oligos are used as the reverse transcription primer. PCR amplification is carried out using miRNA specific forward primer and reverse primer. SYBR Green, an intercalating dsDNA dye, is then used to monitor PCR product formation. (B) DNA-based capture probes immobilized on the microarray are used to capture fluorescently tagged miRNAs. The fluorescent signal is then quantitated and the intensity is related to the relative miRNA expression. (C) Most RNA-sequencing workflows begin by reverse transcribing miRNA into a cDNA library. This is followed by adaptor ligation that allows for immobilization on a substrate that are used to obtain sequencing data.