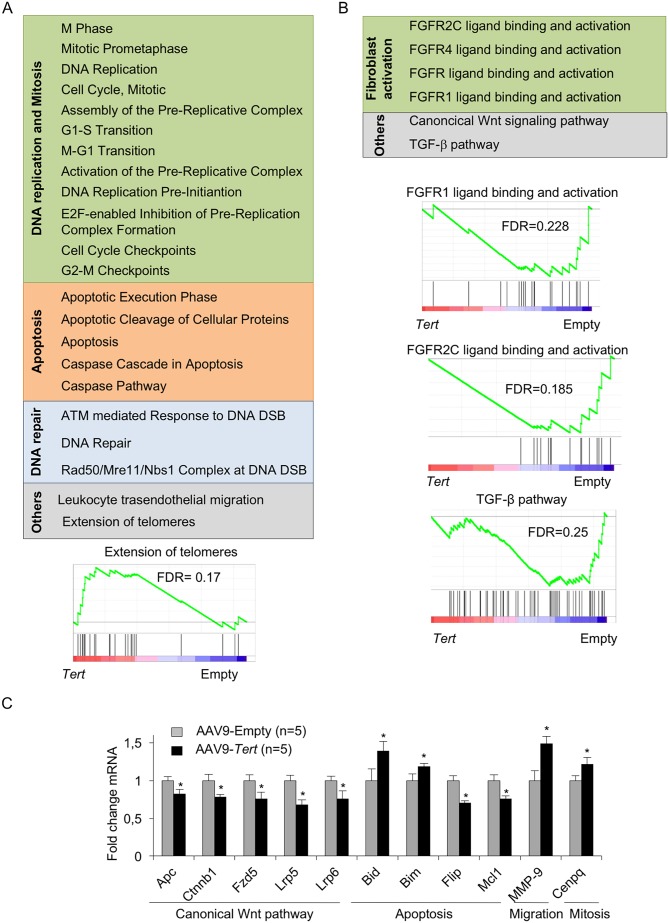
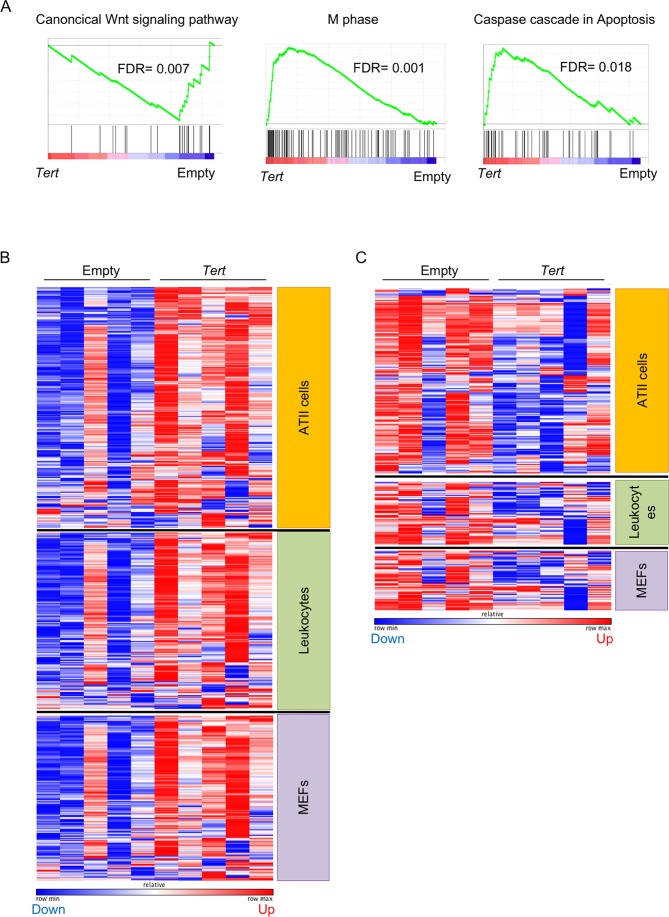
Figure 4. AAV9-Tert treatment induces transcriptional changes in the lungs.
(A–B) Summary table indicating various significantly (FDR < 0.25) upregulated (A) and downregulated (B) pathways in AAV9-Tert compared with AAV9-Empty treated lungs at 8 weeks post-viral treatment. Examples of GSEA plots for the indicated pathways are shown below. Microarray genes were ranked based on the two-tailed t-statistic tests obtained from the AAV9-Tert versus AAV9-Empty by pair-wise comparisons. The red to blue horizontal bar represents the ranked list. Those genes showing higher expression levels for each cohort are located at the edges of the bar (AAV9-Empty; AAV9-Tert). The genes located at the central area of the bar show small differences in gene expression fold changes between both groups. (C) Fold change mRNA expression levels of candidate genes related with canonical Wnt pathway, apoptosis, mitosis and transendothelial migration in AAV9-Tert relative to empty vector. For GSEA Kolmogorov–Smirnoff testing was used for statistical analysis. The FDR is calculated by Benjamini and Hochberg FDR correction. Data represent the mean ± SE of analyzed mice within each group. The number of mice analyzed per group is indicated. T-test was used for q-PCR statistical analysis. *p=0.05.