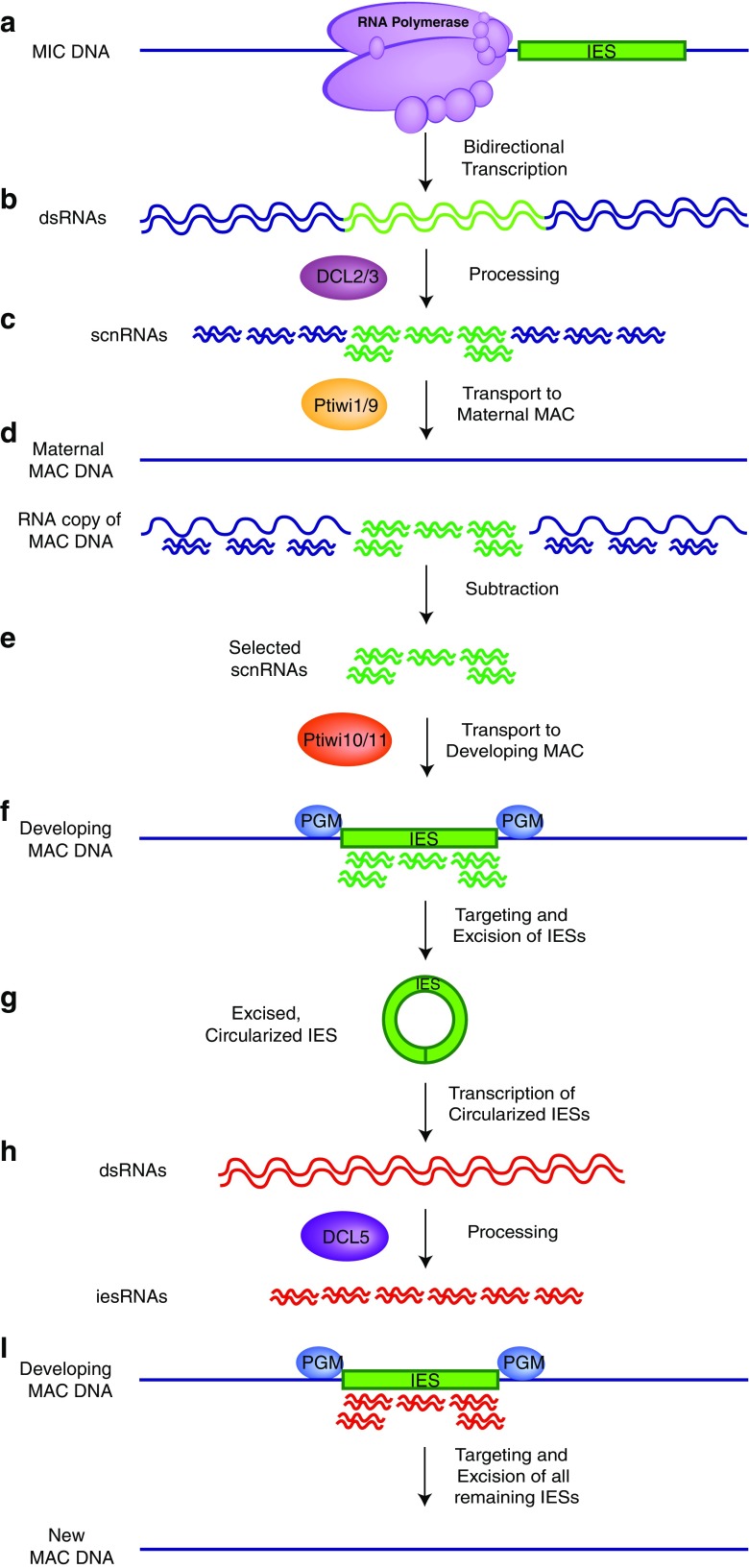
Fig. 1.
Scanning model for DNA elimination in Paramecium tetraurelia. (a) The micronuclear genome is transcribed bidirectionally by an unknown RNA polymerase to produce long double-stranded RNAs. (b) These long dsRNA precursors are processed by the Dicer-like enzymes DCL2/3 to produce 25 nt long scnRNAs. (c) scnRNAs, in association with the PIWI proteins Ptiwi1/9, are transported to the maternal macronucleus (MAC). (d) scnRNAs “scan” the macronuclear genome via interaction with RNA transcripts of somatic DNA. scnRNAs pairing to homologous macronuclear destined sequences (MDSs) are filtered out and degraded, leaving only those corresponding to germline internal eliminated sequences (IESs). (e) Selected scnRNAs, in association with PIWI proteins Ptiwi10/11, are transported to the developing MAC. (f) These scnRNAs target the excision of IESs by the excisase PiggyMac (PGM). (g) Excised IESs circularize, or concatemerize and circularize, and are transcribed into long dsRNAs. (h) These long dsRNA precursors are processed by the Dicer-like enzyme DCL5 to produce 22–31 nt long iesRNAs. (i) iesRNAs ensure the precise and efficient excision of all remaining IESs from the developing macronuclear genome. Development of the new MAC is completed, with the newly formed macronuclear genome matching that of the maternal macronucleus