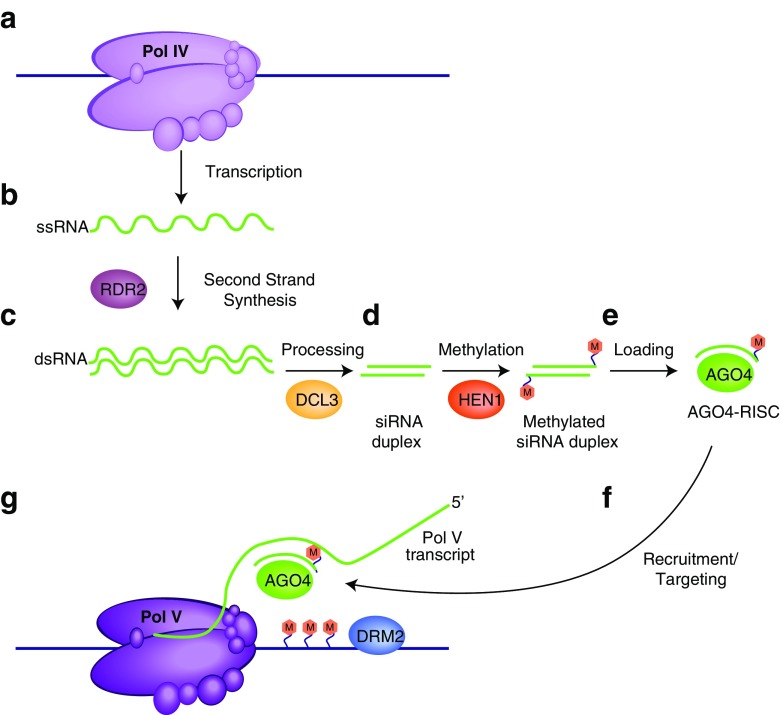
Fig. 2.
RNA-directed DNA methylation (RdDM) in Arabidopsis thaliana. (a) RNA polymerase IV produces single-stranded RNA transcripts that initiate the process of RdDM. (b) These single-stranded RNAs are used as templates for the transcription of a second strand by the RNA-dependent RNA polymerase RDR2. (c) These double-stranded RNA duplexes are cleaved by the Dicer-like enzyme DCL3 to produce 24 nt siRNAs. (d) 24 nt siRNA duplexes are modified by the RNA methylase HEN1, which adds a stabilizing 3′-O-methyl group. (e) A single 24 nt strand of RNA is loaded onto the Argonaute family protein AGO4 to form an active RNA-induced silencing complex (RISC). (f) This sRNA bound RISC complex is then recruited to growing transcripts produced by RNA polymerase V, where direct interaction between AGO4-bound sRNAs and nascent transcripts is thought to occur. (g) De novo cytosine methylation of the corresponding DNA sequence is mediated by the methyltransferase DRM2. This ultimately leads to the removal of active histone marks and the establishment of repressive ones, leading to silencing of specific genomic regions