Fig. 4.
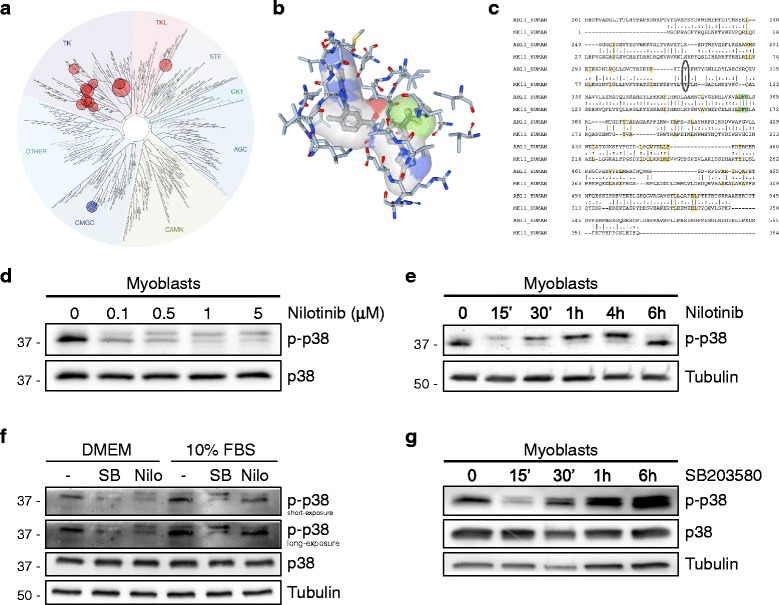
Nilotinib inhibits p38 MAPK in myoblasts. a Analysis of Nilotinib binding across the kinome. Interaction map showing a circular representation of the kinase family tree (KINOMESCAN, https://www.discoverx.com/technologies-platforms/competitive-binding-technology/kinomescan-technology-plataforma). This map describes quantitatively the interaction patterns of Nilotinib using a 100-nM affinity cutoff (Kd). The blue circle labels the interaction of Nilotinib with the p38 MAPK family. b Crystal structure of human MAPK11 (p38β) in complex with Nilotinib. c Alignment of human Abl (upper) and MAPK11 (p38 beta) (lower) aminoacid sequences that are part of the SH-1 kinase domains. Amino acids with “|” are structurally equivalent and identical residues, “:” are structurally equivalent and similar residues, “.” are structurally equivalent, but not similar residues. The threonine gatekeeper residues (boxed), DFG motif (green), and conserved hydrophobic interactions (yellow) are shown. d The myoblasts were treated with increasing concentrations of Nilotinib for 30 min. Cell lysates were collected for Western blotting of phospho-p38 and total p38. e Western blots showing phospho-p38 levels in myoblasts after Nilotinib (5 μM) treatment at different time points. Tubulin was used as loading control. f Representative Western blot of myoblasts that evaluates p38 phosphorylation using SB203580 (SB) (5 μM) p38 inhibitor and Nilotinib. Both treatments were performed for 30 min. Total p38 and tubulin were used as loading controls. g Western blots showing phospho-p38 levels in myoblasts after SB203580 treatment at different time points. Total p38 and tubulin were used as loading controls
