Fig. 6.
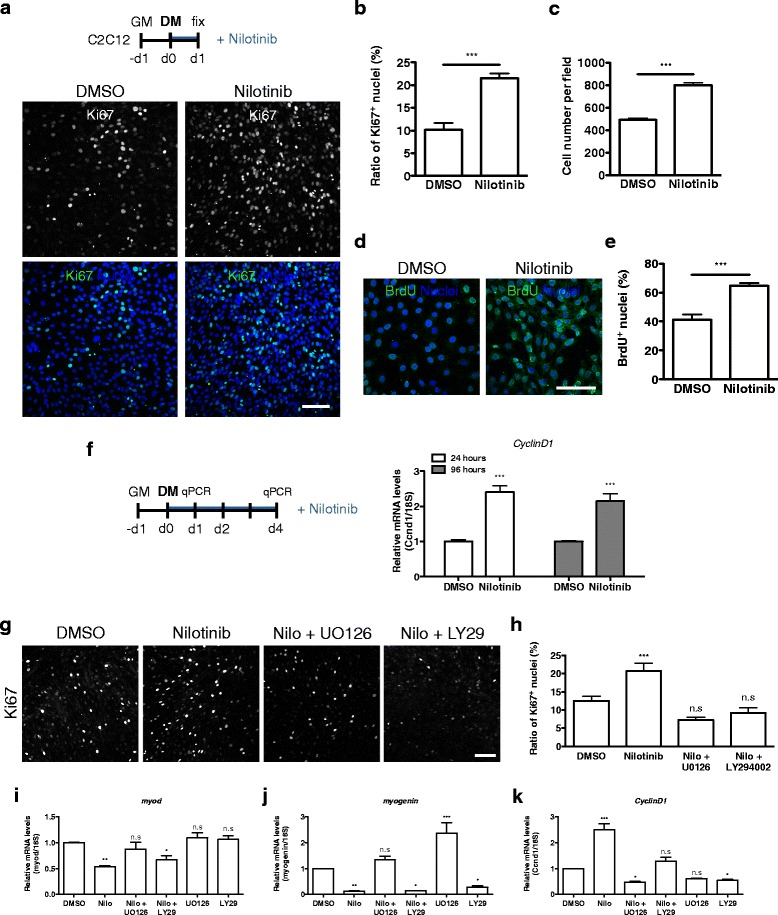
Nilotinib induces myoblast proliferation through ERK1/2 and AKT pathways. a Outline of C2C12 skeletal muscle differentiation and 24 h Nilotinib treatment protocol (upper). Immunofluorescence analyses of the proliferation marker Ki67 (gray upper panels; green lower panels) in C2C12 myoblasts after 24 h of DMSO or Nilotinib (5 μM) in differentiation medium. Hoechst (blue) was used to stain nuclei (lower). Scale bar: 50 μm. b Evaluation of proliferation as the percentage (%) of Ki67-positive cells per field is shown. The values correspond to the mean ± SEM (n = 4). ***P < 0.001; DMSO vs Nilotinib; with two-tailed Student’s t test. c Quantification of total cell number per field is shown. The values correspond to the mean ± SEM. ***P < 0.001; DMSO vs Nilotinib; n = 4; with two-tailed Student’s t test. d Immunofluorescence analysis of C2C12 myoblasts after 24 h in differentiation medium shows nuclear (Hoechst in blue) localization of BrdU proliferation marker. Scale bar: 50 μm. e Quantification of proliferation as the % of BrdU+-cells. The values correspond to the mean ± SEM (n = 3). ***P < 0.001; DMSO vs Nilotinib; with two-tailed Student’s t test. f Outline of C2C12 skeletal muscle differentiation and 24 or 96 h Nilotinib treatment protocol. CyclinD1 expression levels were analyzed by quantitative PCR in C2C12 myoblasts after 24 and 96 h treatments in differentiation medium. Values correspond to the mean ± SEM. n = 4; ***P < 0.001, **P < 0.005, *P < 0.05, n.s not significant. g Representative images of Ki67 immunofluorescence in myoblasts treated for 24 h with DMSO, Nilotinib, Nilo (Nilotinib)+UO126, and Nilo + LY29 (LY294002) in differentiation medium. h Quantification of three independent experiments, evaluating proliferation as the % of Ki67+-cells per field. The values correspond to the mean ± SEM. ***P < 0.001, **P < 0.005, n.s non-significant; n = 3. One-way ANOVA with Bonferroni post-test. i MyoD, j myogenin, and k cyclinD1 expression levels were analyzed by quantitative PCR in C2C12 myoblasts after 24 h of treatments in differentiation medium. Values correspond to the mean ± SEM. ***P < 0.001, **P < 0.005, *P < 0.05, n.s not significant; n = 4. One-way ANOVA with Bonferroni post-test
