Figure 1.
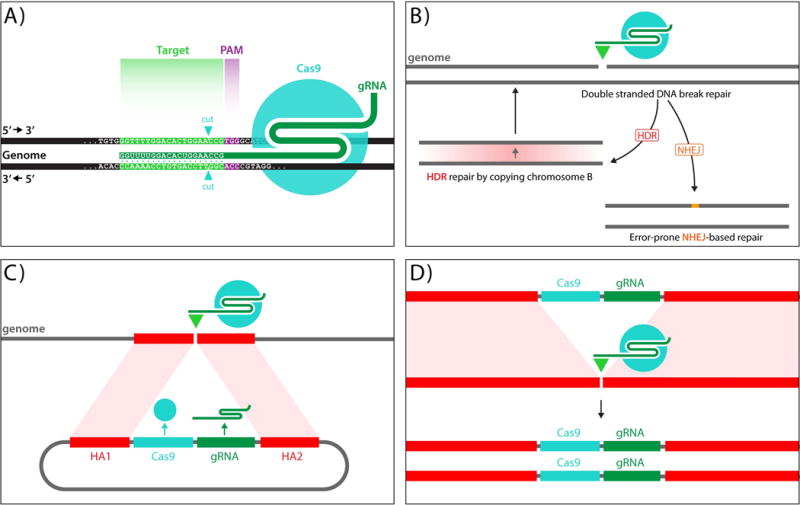
Outline of CRISPR and MCR methods. A) The CRISPR/Cas9 genome editing system consists of two elements, the Cas9 endonuclease, which generates blunt ended double stranded DNA breaks, and a 20 nucleotide guide RNA (gRNA) that binds to Cas9 and targets it to complementary genomic sequences, which in addition must have a so-called PAM sequence (NGG - violet type) recognized by Cas9 that lies immediately 3′ to the 20 nucleotides of gRNA match. B) Double stranded chromosomal breaks caused by targeted cas9/gRNA cleavage can be repaired by either the Rad51-dependent Homology Directed Repair (HDR) pathway, which faithfully copies information from the sister chromosome into the cut site, or the Ku70/80-dependent Non-homologous End-Joining (NHEJ) pathway, which typically results in short insertions/deletions (indels) at the cut site. C,D) MCR mutagenesis scheme: MCR elements (C) consist of three components: 1) a transgene encoding a nuclear targeted form of Cas9 endonuclease, 2) a gRNA directing cleavage to a desired genomic site, and 3) homology arms (HA1 and HA2) from the targeted locus that directly about the gRNA cut site. An injected MCR construct inserts into the genome at the site of gRNA directed cleavage. Once integrated into the genome (D), the MCR element acts on the opposing allele and inserts itself to generate a homozygous insertional mutation.
