Figure 3.
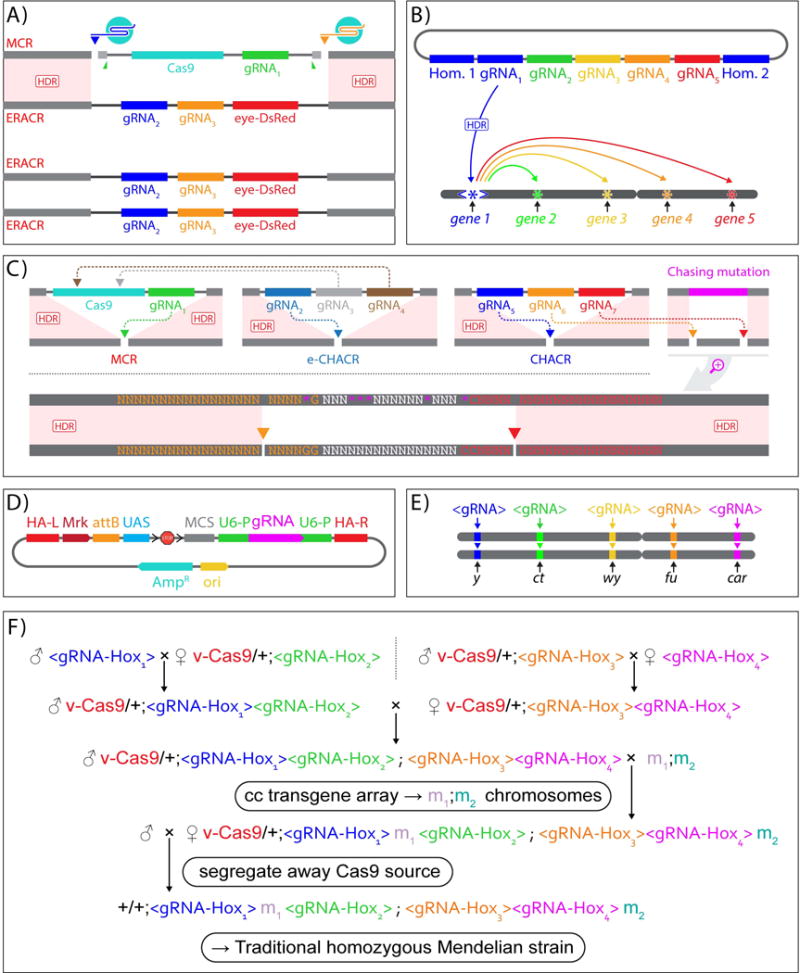
ERACRs, CHACRs, and “copy-cat” <gRNA> constructs: A) ERACRs: “Elements to Reverse the Autocatalytic Chain Reaction” delete MCR elements. In flies carrying both an MCR and an ERACR allele, Cas9 produced by the MCR cuts at sites directed by gRNA-2 and gRNA-3. eye-DsRed = dominant marker. The MCR inserted at a cut site determined by gRNA-1 lying within the deleted segment leading to the ERACR element becoming homozygous. B) CHACRs: “Constructs Hitchhiking on the Autocatalytic Chain Reaction” target other genomic targets. Shown here is an example in which a CHACR serves as a platform to launch an array of gRNAs to diverse targets where they induce standard NHEJ-dependent mutations. C) An MCR element (top left panel) could also be neutralized by CHACR elements used as second-site ERACRs (e-CHACR - inserted at site determined by gRNA2 - top second panel) that carries multiple gRNAs (gRNA3 - teal, gRNA4 - brown) targeting Cas9 in the MCR. Also, CHACRs could be used to drive the spread of unlinked auxiliary elements. Such a CHACR element is shown (top right panel) carrying 3 gRNAs inserted into the cut site of one of these gRNAs (gRNA5 - dark blue), which is in a different location in the genome than the MCR (inserted at a site defined by gRNA1 - green). Thus, like an ERACR, in the presence of an MCR carrying a Cas9 source, the CHACR cuts the opposing chromosome (via cleavage induced by gRNA5) and inserts itself into the resulting DNA gap. In addition, the depicted CHACR carries gRNA6 (orange) and gRNA7 (red), which cut at adjacent sites flanking a edited genomic locus (or existing natural allelic variant - top right panel). The resulting small deletion (region between the gRNA6 and gRNA7 cut sites) will then be repaired via HDR using the edited (pink) sequence. The lower panel shows a magnified view of the top right panel indicating the gene edited residues as pink asterisks and the two cleavage sites for gRNA6 (orange) and gRNA6 (red) relative to the sequences of perfect homology mediating HDR repair. D) “Copy-cat” or cc vectors allow the cloning of transgenes into multiple cloning sites (MCS) as well as matched sets of gRNA(s) flanked by both 5′ (U6p) and 3′ (U6-3′) U6-RNA regulatory elements, and homology arms (HA-L = left, HA-R = right), standard features of cloning vectors such as a bacterial origin of replication (Ori), a gene providing Ampicillin resistance (AmpR), as well as optional use cassettes such as a UAS promoter, an attBϕ31C recombinase donor site allowing for alternative recombinase-driven insertion of the construct into a genomic recipient site (attP), or instead, an attP recipient site to allow recombinase-mediated insertion into the genomically inserted copy-cat element, and an FRT-flanked transcriptional stop cassette (<Stop<). E) cc elements can insert at various loci along a chromosome (D. melanogaster X-chromosome shown as example) which are determined by their particular matched sets of gRNAs and homology arms. In the presence of a cas9 source, these elements will be copied to the sister chromosome, thereby homozygosing the element with the inserted transgene. F) Example of how copy-cat elements could be used in a model vertebrate organism such as a mouse or fish to create a cas9-dependent viable quadruple knock-out of a set of target genes (e.g., redundantly acting Hox gene paralogs). Not shown here for simplicity are various transgene constructs that also could be carried by each of the cc-elements (e.g., CRE/LOX components and fluorescent markers appropriate for expressing and analyzing the ability of a single Hox gene to substitute for the normal sets of genes in a given tissue). These cc elements/mutant alleles could be assembled in two generations. Next, in the maintained presence of cas9, they could be combined with two traditional Mendelian alleles (m1 and m2) by cc-ing the Hox mutant alleles into the mutant background. The source of cas9 then could be removed by segregation, resulting in the complex assembly of mutant alleles and transgenes which would now behave according to standard Mendelian rules.
