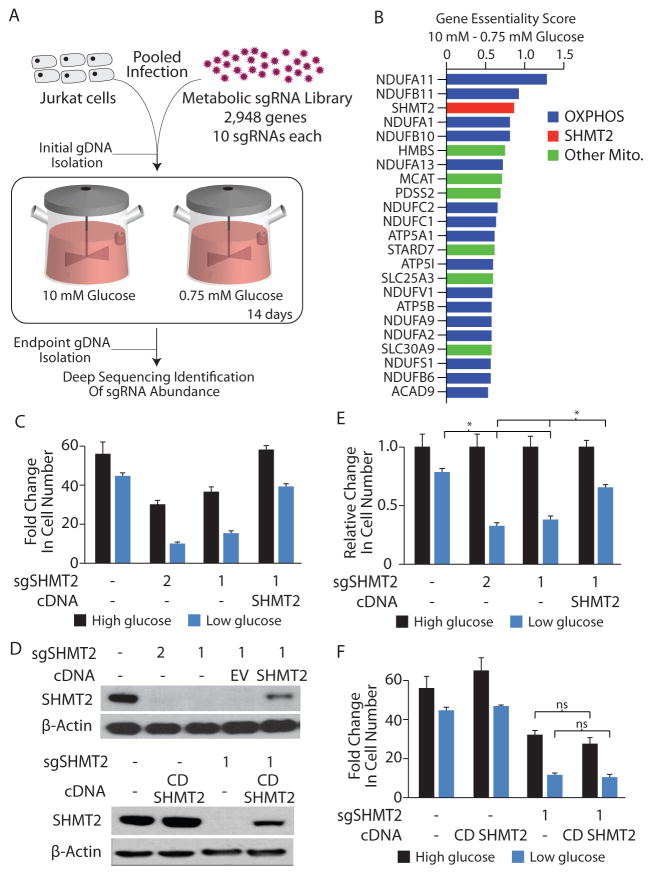
Figure 1. A CRISPR/Cas9 based genetic screen identifies SHMT2 as being differentially required in low glucose conditions.
A, Pooled screening outline. An sgRNA library targeting 2,948 metabolic enzymes and small molecule transporters was transduced into Jurkat T cells followed by culture in Nutrostats set to 10 mM or 0.75 mM glucose for a period of 14 days. Genomic DNA was collected prior to or after the 14 day period and the abundance of sgRNAs determined by deep sequencing. B, Genes exhibiting differential essentiality in 0.75 mM glucose, compared to 10 mM glucose, median Log2 fold change cutoff of 0.5. Genes indicated in blue are components of oxidative phosphorylation complexes, while those in green are also mitochondrially localized. C, Proliferation of Jurkat cells or clones expressing small guide RNAs targeting SHMT2 (sgSHMT2), or with reintroduction of the SHMT2 cDNA. Cells were grown for 5 days in media initially containing 10 mM (high glucose) or 1.5 mM glucose (low glucose). D, Immunoblot for SHMT2 or beta-actin from cell lysates of the lines described in C (above), or expressing a catalytically inactive mutant (K280A) of SHMT2 (CD SHMT2) (below). E, Data from C, normalized to the 10 mM glucose condition for each cell line. F, Proliferation as in C, using the cell lines indicated. * p < 0.05, ns = not significant. Error bars are s.e.m. n=3 for cell proliferation experiments.