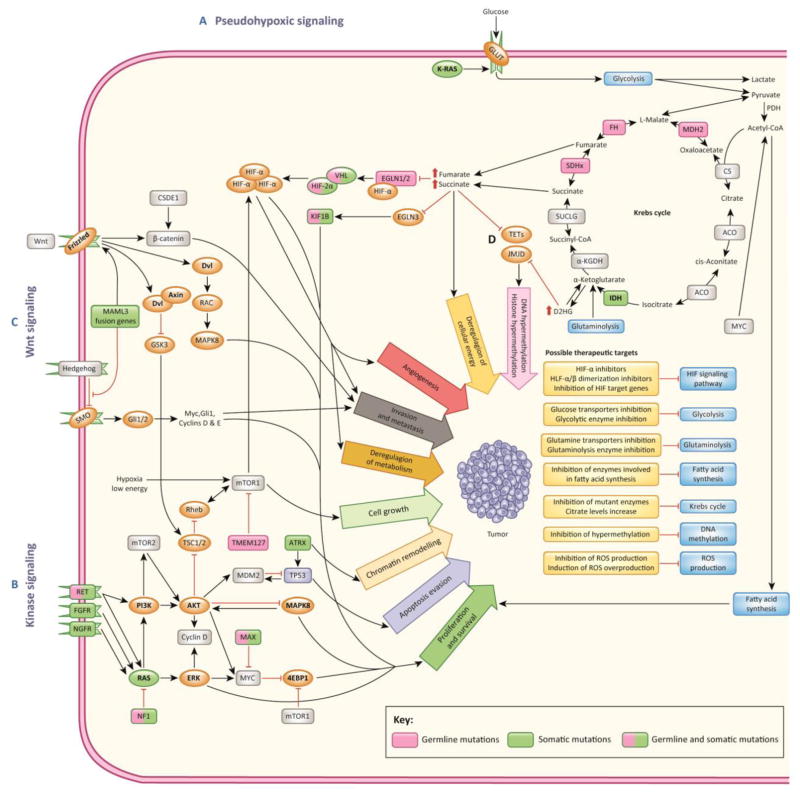
Figure 1. Main signaling clusters activated in PHEO and PGL.
A) Mutations in PHEO/PGL susceptibility genes belonging to the pseudohypoxic signaling cluster result in direct or indirect activation of the hypoxia signaling (HIF) pathway even in the presence of sufficient amount of oxygen (pseudohypoxia). Inactivating mutations in SDHx, SDHAF2, FH, or MDH2 lead to accumulation of Krebs cycle metabolites, such as succinate, fumarate, pyruvate, or glutamine, which is followed by activation of pseudohypoxic signaling and transcription of HIF target genes. VHL, EGLN1/2, and HIF2A genes are directly involved in hypoxic signaling. Mutations in these genes are associated with activation of the HIF signaling pathway. Pseudohypoxic signaling drives cell proliferation, deregulation of cellular energy and metabolism, angiogenesis, invasion and metastasis. B) Kinase signaling cluster includes mutations in genes involved in abnormal activation of kinase signaling, particularly PI3K/AKT, mTOR, and RAS/MAPK pathways. Activation of these pathways is associated with cell proliferation and survival, evasion from apoptosis, and chromatin remodeling. Kinase signaling pathways in PHEO and PGL can be disrupted by mutations in NF1, KIF1B, MAX, RET, TMEM127, H-RAS, and ATRX genes. Rarely, mutations in K-RAS, FGFR genes and fusion genes involving NGFR, BRAF, or NF1 can be found in PHEOs/PGLs. C) Recently, the Wnt signaling pathway has been implied to play role in PHEO/PGL pathogenesis based on the identification of CSDE1 mutations and MAML3 fusion genes [3]. D) Mutations in SDHx and FH genes result in an accumulation of succinate and fumarate and mutated IDH1 activity that results in conversion of α-ketoglutarate to the oncometabolite D2HG. Succinate, fumarate, D2HG, and L2HG function as competitive inhibitors of α-ketoglutarate–dependent dioxygenases. Inhibition of reactions mediated by α-ketoglutarate-dependent dioxygenases leads to DNA and histone hypermethylation, activation of hypoxic responses, and inhibition of collagen maturation and folding.
Abbreviations: 4EBP1, eukaryotic translation initiation factor 4E binding protein 1; α-KGDH, alpha-ketoglutarate dehydrogenase; Acetyl-CoA, acetyl-coenzyme A; ACO, aconitase; AKT, AKT serine/threonine kinase; ATRX, ATRX, chromatin remodeler; BRAF, B-Raf proto-oncogene; CS, citrate synthase; CSDE1, cold shock domain containing E1; D2HG, D-2-hydroxyglutarate; Dvl, Dishevelled segment polarity protein; EGLN1/2/3, egl-9 prolyl hydroxylase 1, 2, 3; ERK, mitogen-activated protein kinase 1; FGFR, fibrablast growth factor receptor; FH, fumarate hydratase; Gli1/2, GLI family zinc finger 1, 2; GLUT, glucose transporter; GSK3, glycogen synthase kinase 3; HIF, hypoxia-inducible factor; HIF2A, hypoxia-inducible factor 2 alpha; H-RAS, HRas proto-oncogene, GTPase; IDH1/2, isocitrate dehydrogenase 1, 2; JMJD, Jumoni C domain-containing histone lysine demethylases; KIF1B, kinesin family member 1B; K-RAS, KRAS proto-oncogene, GTPase; L2HG, L-2-hydroxyglutarate; MAPK, mitogen activated kinase; MAML3, mastermind like transcriptional coactivator 3; MAX, MYC associated factor X; MDH2, malate dehydrogenase 2; MDM2, MDM2 proto-oncogene; mTOR, mechanistic target of rapamycin; MYC, MYC proto-oncogene; NF1, neurofibromin 1; NGFR, nerve growth factor receptor; PDH, pyruvate dehydrogenase; PHD1/2, egl-9 family hypoxia inducible factor 2, 1; PHEO/PGL, pheochromocytoma/paraganglioma; PI3K, phosphatidylinositol-3-kinase; RAC, Rho guanine nucleotide exchange factors; RAS, RAS type GTPase family; RET, RET proto-oncogene; Rheb, Ras homolog enriched in brain; ROS, reactive oxygen species; SDHx, succinate dehydrogenase subunits A, B, C, D, SDHAF2, succinate dehydrogenase complex assembly factor 2; SMO, Smoothened, frizzled class receptor; SUCLG, succinyl-coenzyme A ligase; TETs, tet methylcytosine dioxygenases; TMEM127, transmembrane protein 127; TP53, tumor protein 53; TSC1/2, TSC complex subunit 1, 2; VHL, von Hippel-Lindau tumor suppressor,