Significance
In plants, RNA interference (RNAi) is the main antiviral defense mechanism. RNAi moves cell-to-cell through cytoplasmic channels called plasmodesmata, spreading ahead of the viral infection to immunize tissues before arrival of the virus. How this movement is regulated has been elusive. Here, we identify a plant protein, BAM1, localized at plasmodesmata and promoting RNAi spread; both BAM1 and its closest homolog, BAM2, are required for this process. In agreement with their role in promoting RNAi movement, we found that BAM1 and BAM2 are targeted by a viral effector, which acts as a suppressor of RNAi spread. Our work offers insight into the regulation of cell-to-cell spread of RNAi and provides another example of the arms race between pathogens and hosts.
Keywords: receptor-like kinase, plasmodesmata, RNAi, BAM1, Geminivirus
Abstract
RNA interference (RNAi) in plants can move from cell to cell, allowing for systemic spread of an antiviral immune response. How this cell-to-cell spread of silencing is regulated is currently unknown. Here, we describe that the C4 protein from Tomato yellow leaf curl virus can inhibit the intercellular spread of RNAi. Using this viral protein as a probe, we have identified the receptor-like kinase (RLK) BARELY ANY MERISTEM 1 (BAM1) as a positive regulator of the cell-to-cell movement of RNAi, and determined that BAM1 and its closest homolog, BAM2, play a redundant role in this process. C4 interacts with the intracellular domain of BAM1 and BAM2 at the plasma membrane and plasmodesmata, the cytoplasmic connections between plant cells, interfering with the function of these RLKs in the cell-to-cell spread of RNAi. Our results identify BAM1 as an element required for the cell-to-cell spread of RNAi and highlight that signaling components have been coopted to play multiple functions in plants.
RNA interference (RNAi) mediated by small interfering RNA (siRNA) is considered the main antiviral defense mechanism in plants. RNAi relies on the production of virus-derived siRNA (vsiRNA) by RNaseIII Dicer-like proteins, mainly DCL2 and its surrogate DCL4; vsiRNAs are then loaded into argonaute (AGO) proteins AGO1- and AGO2-containing complexes to target viral RNA for cleavage (1). siRNAs can move from cell to cell (2, 3), presumably symplastically through plasmodesmata (PD), and it has been proposed that vsiRNAs can move ahead of the front of the infection, leading to spread of silencing and immunizing plant tissues before the arrival of the virus. However, how cell-to-cell movement of siRNAs/vsiRNAs occurs remains elusive, and efforts to identify plant proteins specifically involved in this process through forward genetics have not been fruitful (3, 4).
To counter antiviral RNAi, viruses have evolved viral suppressors of RNA silencing (VSR). All plant viruses described to date encode at least one VSR, supporting the notion that active suppression of RNAi is a conditio sine qua non for infectivity. Although independently evolved VSRs have been shown to target different steps of the RNAi pathway (5), direct interference of a VSR with cell-to-cell movement of the silencing signal has not yet been characterized.
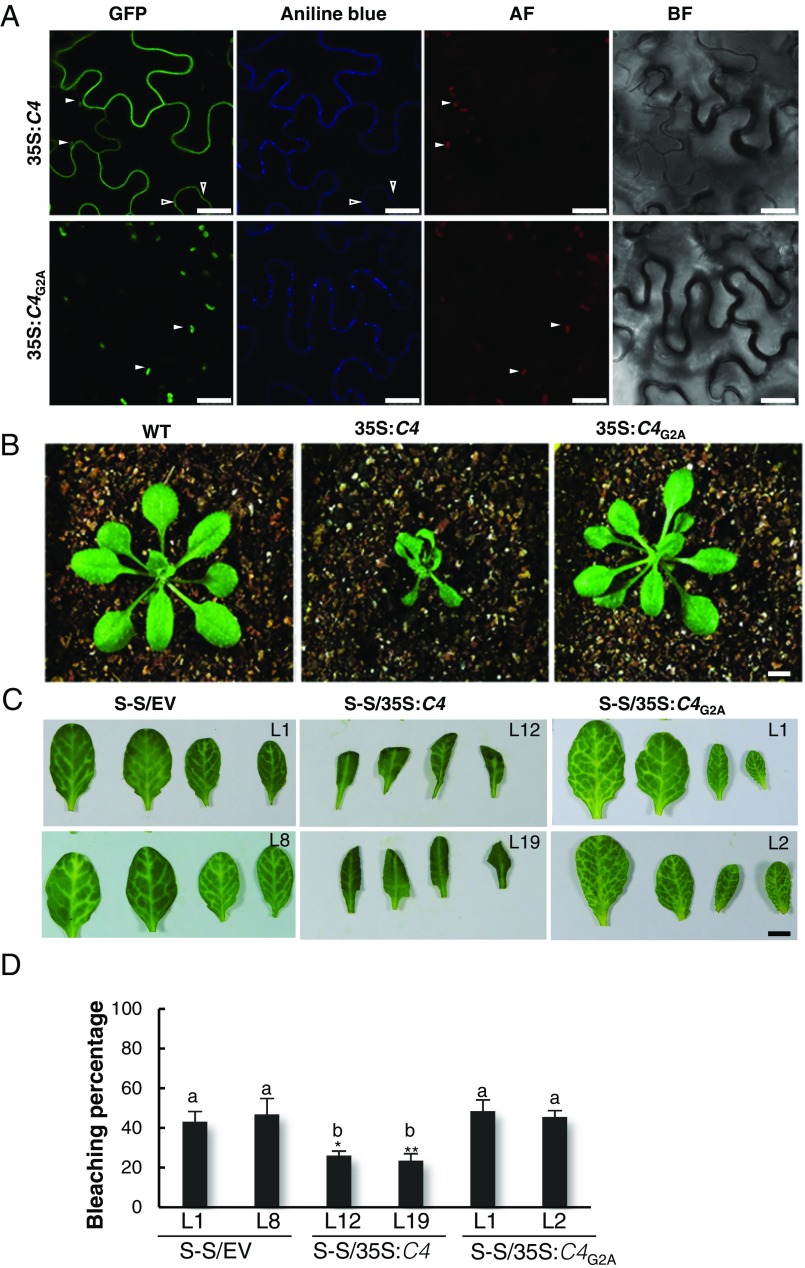
The C4 protein from Tomato yellow leaf curl virus (TYLCV; family Geminiviridae) can delay the systemic spread of silencing in transgenic Nicotiana benthamiana plants, but is not a local suppressor of RNAi (6). We therefore set out to assess whether C4 could interfere with cell-to-cell movement of silencing. Interestingly, we found that C4 localizes to different compartments in the plant cell, mainly the plasma membrane (PM), plasmodesmata (PD), and chloroplasts; PM/PD localization depends on the presence of a myristoylation motif at the N terminus of the protein (Fig. 1A and SI Appendix, Fig. S1). A mutant virus expressing a nonmyristoylable version of C4 (C4G2A), which localizes to chloroplasts exclusively, displays severely compromised infectivity in tomato (SI Appendix, Fig. S2), suggesting that PM/PD localization is required for full infectivity. When expressed constitutively in Arabidopsis, C4 causes strong developmental alterations, which also require myristoylation (Fig. 1B and SI Appendix, Fig. S3).
Fig. 1.
Plasma membrane/plasmodesmal C4 suppresses cell-to-cell spread of RNAi. (A) Subcellular localization of C4-GFP or the nonmyristoylable mutant C4G2A-GFP upon transient expression in Nicotiana benthamiana leaves. BF, Bright field; AF, Autofluorescence. Empty arrowheads indicate plasmodesmata; filled arrowheads indicate chloroplasts. (Scale bar, 25 µm.) (B) Transgenic plants expressing C4 or the nonmyristoylable mutant C4G2A at 5 wk postgermination. (Scale bar, 0.5 cm.) (C) Leaves of 4-wk-old transgenic SUC:SUL plants expressing C4 (S-S/35S:C4), the nonmyristoylable mutant C4G2A (S-S/35S:C4G2A), or transformed with the empty vector (S-S/EV). Each set of four leaves comes from one T2 plant from an independent line. (Scale bar, 0.5 cm.) (D) Quantification of the bleaching percentage of the leaves in B. Error bars represent SE; bars with the same letter are not significantly different (P = 0.05) according to Dunnet’s multiple comparison test.
To investigate whether C4 affects the cell-to-cell spread of RNAi, we expressed this protein in the SUC:SUL silencing reporter system (2). SUC:SUL transgenic Arabidopsis plants express an inverted repeat of the endogenous SULFUR (SUL) gene (At4g18480) in phloem companion cells, which leads to the production of 21- and 24-nt siRNAs against SUL and the ensuing RNAi, resulting in a chlorotic phenotype of the silenced cells. Cell-to-cell movement of 21-nt siRNAs causes the spread of silencing to 10–15 cells beyond the vasculature (ref. 3, Fig. 1C). Expression of wild-type C4, but not of the nonmyristoylable mutant C4G2A, greatly diminishes the spread of the silencing phenotype, suggesting that PM/PD-localized C4 may interfere with the cell-to-cell movement of silencing from the vasculature (Fig. 1C).
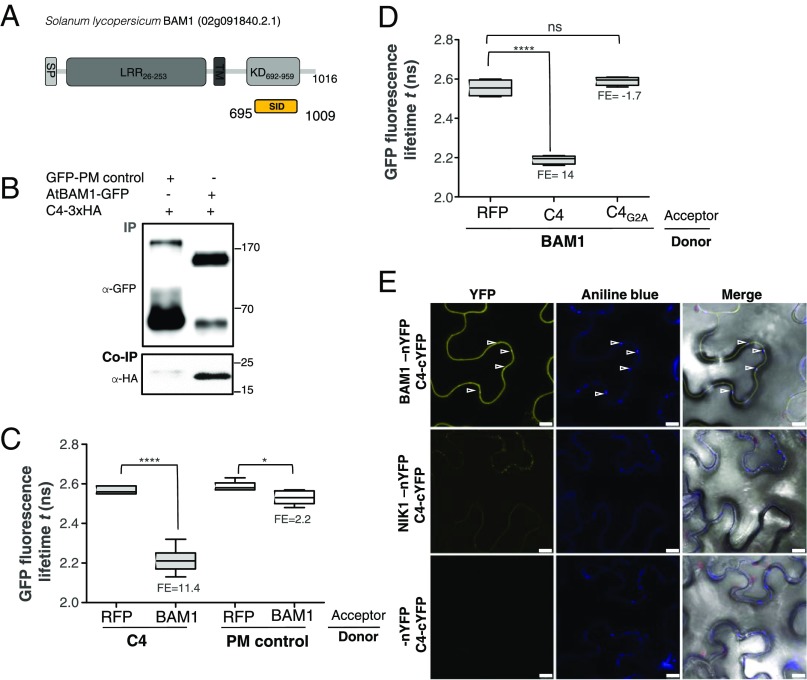
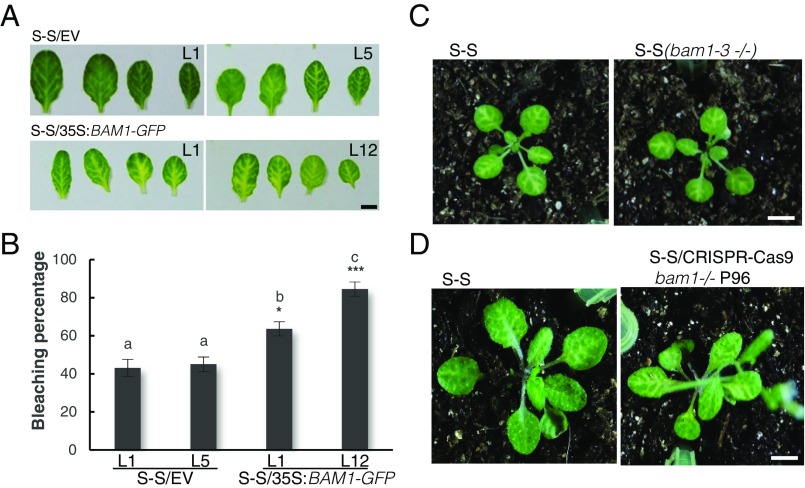
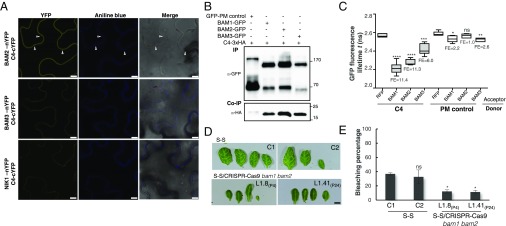
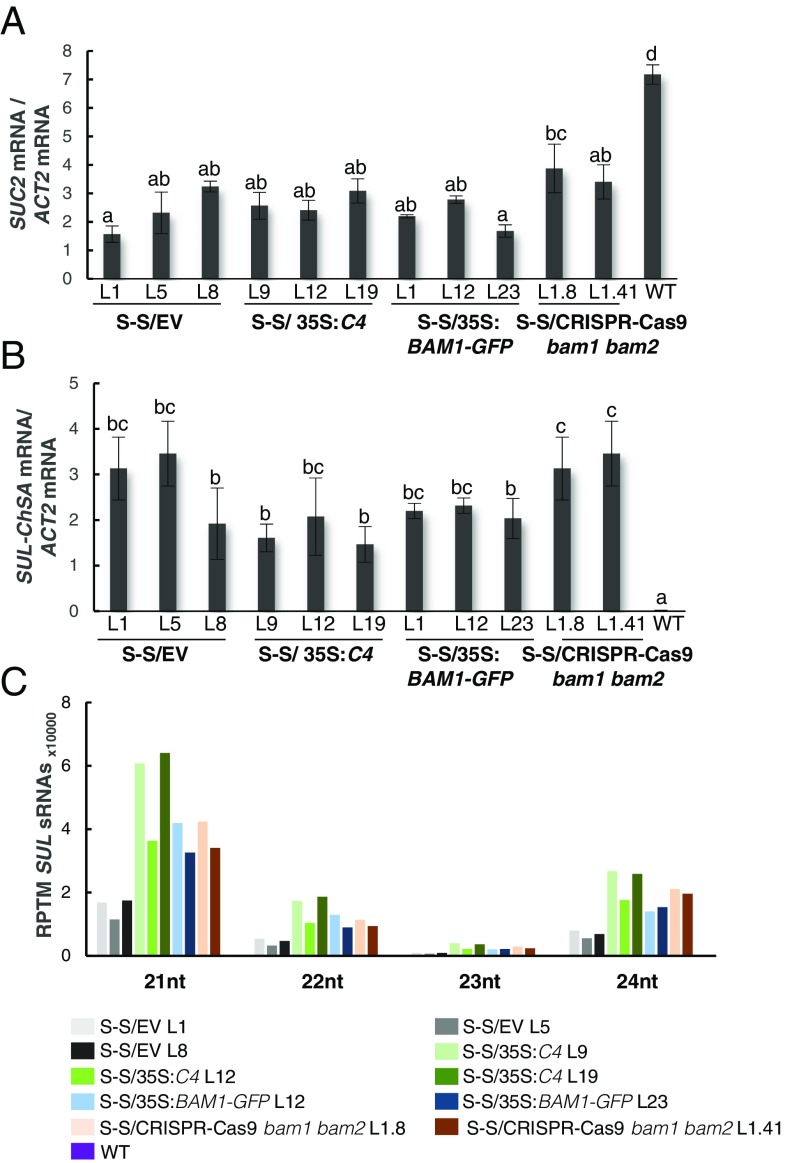
With the aim of uncovering the molecular mechanism of the C4-mediated suppression of cell-to-cell spread of silencing, we searched for interacting partners of C4 through yeast two-hybrid screening of a TYLCV-infected tomato cDNA library and affinity purification followed by mass spectrometry analyses (AP-MS) upon transient expression of C4-GFP in N. benthamiana. Interestingly, these independent approaches both identified the receptor-like kinase (RLK) BARELY ANY MERISTEM 1 (BAM1) as an interactor of C4; the interaction with C4 occurs through the kinase domain of BAM1, as shown by mapping of this interaction in yeast (Fig. 2A). The interaction between C4 and BAM1 from Arabidopsis was confirmed using coimmunoprecipitation (Co-IP), Fӧrster resonance energy transfer–fluorescence lifetime imaging (FRET-FLIM), bimolecular fluorescence complementation (BiFC), and gel filtration chromatography, and it requires PM/PD localization of C4 (Fig. 2 and SI Appendix, Fig. S4). The interaction of C4 with BAM1 from tomato, natural host of TYLCV, was confirmed by BiFC and FRET-FLIM (SI Appendix, Fig. S5). Interestingly, BAM1 is strongly expressed in the vasculature, to which many viruses, including TYLCV, are restricted (ref. 7, SI Appendix, Fig. S6). BiFC assays revealed that C4 interacts with BAM1 at PM and PD (Fig. 2E), where both proteins colocalize (SI Appendix, Fig. S7), thus it is conceivable that BAM1 participates in the cell-to-cell movement of silencing and is targeted by C4. To test if BAM1 is indeed involved in this process, we generated transgenic SUC:SUL plants overexpressing tagged and untagged versions of the endogenous protein (SUC:SUL/35S:BAM1, SUC:SUL/35S:BAM1-GFP, SUC:SUL/35S:BAM1-FLAG) as well as tomato BAM1 (SUC-SUL/35S:SlBAM1-GFP). In all cases, overexpression of BAM1 resulted in an extended spread of silencing from the vasculature (Fig. 3A and SI Appendix, Fig. S8), suggesting that BAM1 promotes cell-to-cell movement of the silencing signal. Kinase activity does not seem to be required for this function, since overexpression of a BAM1D820N mutant, mutated in the catalytic aspartic acid and devoid of autophosphorylation activity, produces a promotion of the spread of silencing indistinguishable from that by the wild-type protein (SI Appendix, Fig. S9). A mutation in the BAM1 gene alone did not affect the silencing phenotype of the SUC:SUL plants (Fig. 3 C and D and SI Appendix, Fig. S10), which suggests functional redundancy. In Arabidopsis, BAM1 has two close homologs, named BAM2 and BAM3; the expression pattern of BAM2, but not of BAM3, seems to overlap with that of BAM1 in roots (SI Appendix, Fig. S6). The kinase domains of BAM1 and BAM2 are 94% identical at the protein level (SI Appendix, Fig. S11), whereas those of BAM1 and BAM3 are 73% identical. Consistent with this, C4 can also interact with BAM2 and, more weakly, with BAM3, as observed in co-IP, BiFC, and FRET-FLIM assays (Fig. 4 A–C). Strikingly, simultaneous mutation of BAM1 and BAM2 in SUC:SUL plants by CRISPR-Cas9–mediated genome editing (8) (SUC:SUL/CRISPR-Cas9 bam1 bam2 plants; SI Appendix, Fig. S12 and Table S1) markedly diminished the spread of silencing from the vasculature, strongly resembling the effect of C4 expression (Fig. 4 D and E and SI Appendix, Figs. S13 and S14). Similar to C4 transgenic plants, silencing of SUL can still be observed around the midvein in SUC:SUL/CRISPR-Cas9 bam1 bam2 plants, but spread of silencing from secondary veins is basically abolished. It has been reported that a bam1 bam2 double mutant shows defects in leaf vein patterning (9); however, these defects do not seem to underlie the reduction in SUL silencing spread (SI Appendix, Fig. S14). Moreover, the expression of the endogenous SUC2 gene or the SUC:SUL transgene is not significantly different between SUC:SUL/EV (control) and SUC:SUL/C4, SUC:SUL/35S:BAM1-GFP, or SUC:SUL/CRISPR-Cas9 bam1 bam2 (Fig. 5 A and B). Quantification of the SUL sRNAs in these transgenic lines by sRNA sequencing demonstrates that differences in sRNA biogenesis or accumulation cannot account for the observed phenotypes (Fig. 5C and SI Appendix, Fig. S13B); this notion is further confirmed by Northern blot (SI Appendix, Fig. S13C). Therefore, we conclude that BAM1 and BAM2 redundantly promote cell-to-cell movement of silencing from the vasculature, and C4 targets both homologs to inhibit their activity in this process.
Fig. 2.
C4 interacts with the receptor-like kinase BAM1. (A) Schematic representation of the receptor-like kinase BAM1 from tomato (Solanum lycopersicum) isolated in the yeast two-hybrid screen as an interactor of C4. The signal peptide (SP), leucine-rich repeats (LRR), transmembrane domain (TM), and kinase domain (KD) are shown; numbers indicate the beginning and end of each domain, in amino acids. The selected interaction domain (SID) is the minimal fragment found to interact with C4 in this screen. (B) Coimmunoprecipitation of BAM1-GFP from Arabidopsis with C4-3xHA upon transient coexpression in N. benthamiana leaves. Numbers on the right indicate molecular weight. (C) Interaction between C4 and BAM1 by FRET-FLIM upon transient coexpression in N. benthamiana leaves. The membrane protein NP_564431 (NCBI) is used as a negative control (PM control). FE, FRET efficiency. Asterisks indicate a statistically significant difference (****, P value < 0.0001; *P value < 0.05), according to a Student’s t test. (D) Interaction between BAM1 and C4 and BAM1 and C4G2A by FRET-FLIM upon transient coexpression in N. benthamiana leaves. FE, FRET efficiency. Asterisks indicate a statistically significant difference (****, P value < 0.0001), according to a Student’s t test. (E) Interaction between C4 and BAM1 by BiFC upon transient coexpression in N. benthamiana leaves. The receptor-like kinase NIK1 is used as a negative control. Arrowheads indicate plasmodesmata. (Scale bar, 10 µm.)
Fig. 3.
BAM1 promotes the cell-to-cell spread of RNAi. (A) Leaves of 4-wk-old transgenic SUC:SUL plants overexpressing BAM1-GFP (S-S/35S:BAM1-GFP) or transformed with the empty vector (S-S/EV). Each set of leaves comes from one T2 plant from an independent line. (B) Quantification of the bleaching percentage of the leaves in A. Error bars represent SE; bars with the same letters are not significantly different (P = 0.05) according to Dunnet’s multiple comparison test. (C and D) Phenotype of SUC:SUL plants mutated in bam1. (C) A bam1-3 mutant homozygous for the SUC:SUL transgene (F3); (D) A bam1 mutant obtained by genome editing with CRISPR-Cas9 (for details, see SI Appendix, Fig. S12 and Table S1). (Scale bar, 0.5 cm.)
Fig. 4.
BAM1 and its homolog BAM2 are required for the cell-to-cell spread of RNAi. (A) Interaction between C4 and BAM2 by BiFC upon transient coexpression in N. benthamiana leaves. The receptor-like kinase NIK1 is used as negative control. Arrowheads indicate plasmodesmata. (Scale bar, 10 µm.) (B) Coimmunoprecipitation of BAM1-GFP, BAM2-GFP, and BAM3-GFP with C4-3xHA upon transient coexpression in N. benthamiana leaves. Numbers on the right indicate molecular weight. (C) Interaction between C4 and BAM1, BAM2, and BAM3 by FRET-FLIM upon transient coexpression in N. benthamiana leaves. The membrane protein NP_564431 (NCBI) is used as a negative control (PM control). FE, FRET efficiency. Asterisks indicate a statistically significant difference (****, P value < 0.0001; ***, P value < 0.005; **P value < 0.01; *P value < 0.05), according to a Student’s t test; ns, not significant. (D) Leaves of 4-wk-old transgenic SUC:SUL plants mutated in bam1 and bam2 obtained by genome editing with CRISPR-Cas9 (for details, see SI Appendix, Fig. S12 and Table S1). Each set of leaves comes from one T3 plant from an independent line. (Scale bar, 0.5 cm.) (E) Quantification of the bleaching percentage of the leaves in D. Error bars represent SE. Asterisks indicate a statistically significant difference (*P value < 0.05), according to a Student’s t test; ns, not significant.
Fig. 5.
Expression of the endogenous SUC2 gene and the SUC:SUL transgene and accumulation of SUL sRNAs in SUC:SUL transgenic plants expressing C4, overexpressing BAM1, or mutated in BAM1 and BAM2. (A) Expression of the endogenous SUC2 gene as measured by qPCR. Bars represent the mean of nine leaves from three 4-wk-old plants (three leaves/plant). (B) Expression of the SUC:SUL transgene as measured by qPCR. Bars represent the mean of nine leaves from three 4-wk-old plants (three leaves/plant). Error bars represent SE; bars with the same letters are not significantly different (P = 0.05) according to Dunnet’s multiple comparison test. Representative leaves used in A and B are shown in SI Appendix, Fig. S13A. (C) Quantification of SUL sRNAs. Total RNA was extracted from a pool of 10 2-wk-old seedlings (depicted in SI Appendix, Fig. S13B). nt, nucleotides; RPTM, reads per 10 million.
Although other functions of BAM1/BAM2 may also be inhibited by C4, as suggested by similarities in developmental defects of bam1 bam2 mutants and C4-expressing transgenic plants (ref. 9, SI Appendix, Fig. S15), C4 does not impair all activities of BAM1, since C4-expressing transgenic plants display a wild-type–like response to CLV3p in root elongation assays, which is BAM1-dependent (ref. 10, SI Appendix, Fig. S16). Taken together, our results show that C4 targets BAM1/BAM2 and suppresses a subset of the functions of these RLKs, including the promotion of the cell-to-cell spread of silencing from the vasculature. This effect of BAM1/BAM2 on intercellular spread of RNAi is specific, since general plasmodesmal conductance is not affected by expression of C4 or overexpression of BAM1 (SI Appendix, Fig. S17).
Using a viral protein as a probe, we have identified the PM/PD-localized RLK BAM1 as a positive regulator of the cell-to-cell movement of RNAi from the vasculature and determined that BAM1 and BAM2 are functionally redundant in this process. Since pathogen effectors have evolved to overcome genetic redundancy, their use to investigate cellular processes is a valuable complement to traditional forward genetic screens.
BAM1/BAM2 had been previously characterized with respect to their role in development (10–12). Our work identifies a function of these RLKs in the cell-to-cell spread of RNAi; however, the underlying molecular mechanism is still elusive. The finding that kinase activity seems to be expendable for the BAM1-mediated promotion of cell-to-cell spread of silencing from the vasculature suggests that these RLKs may have a scaffolding rather than an enzymatic function in this process. Whether BAM1/BAM2 can bind siRNA-binding proteins, such as AGO proteins, or siRNA molecules directly remains to be determined. Strikingly, C4 seems to inhibit certain functions of BAM1/BAM2, including the promotion of intercellular RNAi spread, but not all: this differential targeting may have evolved to avoid detrimental pleiotropic effects of unspecifically suppressing every role of a multifunctional target. The specificity of the C4-mediated inhibition of BAM1 functions may be determined by subcellular localization and composition of the potential BAM1-containing protein complexes involved in the different signaling pathways.
Another open question is whether BAM1/BAM2 also participate in the cell-to-cell movement of other sRNAs, such as miRNAs or tasiRNAs, and if C4 targets these non–cell-autonomous processes as well, dependent or independently of BAM1/BAM2. Mobile sRNAs have been suggested to act as morphogens, determining the proper development of xylem and the establishment of leaf polarity (13–16); the potential role of BAM1/BAM2 and the effect of C4 in these processes is currently under investigation.
Materials and Methods
Plant Material.
All Arabidopsis mutants and transgenic plants used in this work are in the Col-0 background. The bam1-3 mutant is described in ref. 10. The SUC:SUL transgenic line is described in ref. 2. To generate the 35S:C4, 35S:C4G2A, pBAM1:YFP-NLS, and 35S:BAM1 lines, wild-type Arabidopsis plants were transformed with pGWB2-C4, pGWB2-C4G2A, pBAM1:YFP-NLS, and pGWB2-BAM1, respectively (see Plasmids and Cloning). To generate the SUC:SUL/35S:C4, SUC:SUL/35S:C4G2A, SUC:SUL/35S:BAM1, SUC:SUL/35S:BAM1-GFP, SUC:SUL/35S:BAM1-FLAG, and SUC:SUL/35S:SlBAM1-GFP lines, SUC:SUL plants were transformed with the corresponding plasmids (pGWB2-C4, pGWB2-C4G2A, pGWB2-BAM1, pGWB505-BAM1, pGWB511-BAM1, and pGWB505-SlBAM1, respectively) (see Plasmids and Cloning) using the floral dipping method. To generate the bam1-3/SUC:SUL plants, SUC:SUL plants were crossed to bam1-3 plants, and homozygous individuals for both mutation and transgene were identified and phenotyped in F3.
To generate the bam1 bam2/SUC:SUL lines, SUC:SUL plants were transformed with CRISPR-Cas9 constructs to target BAM1 and BAM2 (see Plasmids and Cloning). Mutation of BAM1 and BAM2 was confirmed in T1 by sequencing, and those plants carrying mutations were selected for further characterization in subsequent generations. Homozygous or biallelic mutant plants were identified by sequencing and phenotyped in T2 and T3.
The tomato cultivar used in this work for viral infection assays and cloning of SlBAM1 is Money maker.
Coimmunoprecipitation and Protein Analysis.
Protein extraction, coimmunoprecipitation, and protein analysis were performed as described in ref. 17, with minor modifications. To efficiently extract membrane proteins, 2% of Nonidet P-40 was used in the protein extraction buffer. The antibodies used are as follows: anti-GFP (Abiocode M0802-3a), anti-HA (Santa-Cruz sc-7392), anti-Rabbit IgG (Sigma A0545), and anti-Mouse IgG (Sigma A2554).
Confocal Imaging.
N. benthamiana plants were agroinfiltrated with clones to express C4-GFP, C4-RFP, C4G2A-GFP, and BAM1-GFP, and samples were imaged 2 d later on a Leica TCS SMD FLCS point scanning confocal microscope using the preset settings for GFP with Ex: 489 nm, Em: 500–550 nm, and for RFP with Ex: 554 nm, Em: 580–630 nm. For callose deposition at plasmodesmata, samples were stained with a solution of 0.05% (wt/vol) aniline blue in water by infiltration into leaves 30 min before imaging with Ex: 405 nm, Em: 448–525 nm using sequential scanning.
Bimolecular Fluorescent Complementation.
The plasmids used for BiFC are described in ref. 18 (see Plasmids and Cloning). N. benthamiana plants were agroinfiltrated with clones to express the corresponding proteins, and samples were imaged 2 d later on a Leica TCS SP8 confocal microscope, using the preset settings for YFP with Ex: 514 nm, Em: 525–575 nm.
FRET-FLIM Imaging.
For FRET-FLIM experiments, donor proteins (fused to GFP) were expressed from vectors pGWB5 or pGWB606, and acceptor proteins (fused to RFP) were expressed from vector pB7WRG2.0. FRET-FLIM experiments were performed on a Leica TCS SMD FLCS confocal microscope excitation with WLL (white light laser) and emission collected by a SMD SPAD (single photon-sensitive avalanche photodiodes) detector. Leaf discs of N. benthamiana plants transiently coexpressing donor and acceptor, as indicated in the figures, were visualized 2 d after agroinfiltration. Accumulation of the GFP- and RFP-tagged proteins was estimated before measuring lifetime. The tunable WLL set at 489 nm with a pulsed frequency of 40 MHz was used for excitation, and emission was detected using SMD GFP/RFP Filter Cube (with GFP: 500–550 nm). The fluorescence lifetime shown in the figures corresponding to the average fluorescence lifetime of the donor (τ) was collected and analyzed by PicoQuant SymphoTime software. Lifetime is normally amplitude-weighted mean value using the data from the single (GFP-fused donor protein only or GFP-fused donor protein with free RFP acceptor or with noninteracting RFP-fused acceptor protein) or biexponential fit (GFP-fused donor protein interacting with RFP-fused acceptor protein). Mean lifetimes are presented as means ± SD based on more than 10 cells from at least three independent experiments. FRET efficiency was calculated according to the formula E = 1 − τDA/τD, where τDA is the average lifetime of the donor in the presence of the acceptor and τD is the average lifetime of the donor in the absence of the acceptor.
Plasmids and Cloning.
Plasmids and primers used for cloning are summarized in SI Appendix, Table S2. The TYLCV clone used as template is AJ489258 (GenBank).
Vectors from the pGWB and ImpGWB series were kindly provided by Tsuyoshi Nakagawa (19, 20). pB7RWG2.0 and pBGYN are described in refs. 21 and 22, respectively. The vectors used for CRISPR-Cas9–mediated genome editing are described in ref. 8, and the sequences to target BAM1 and BAM2 were TCTCCGGTCTCAACCTCTC and CTCACCTCTCCTGACCTCA, respectively. The vector used for expression in Escherichia coli is pET21b (Novagen).
Quantitative RT-PCR.
Quantitative RT-PCR was performed as described in ref. 17. Primers used are as follows SlBAM1: CTTGCCCTGAAAACTGCCAT and CGTGACACCATTCCACGTAC (primer pair efficiency: 90%); SUC2: TCTAGCCATTGTCGTCCCTC and CGCCAATACACCACTTACCG (primer pair efficiency: 99%); SUL-ChsA: CCGAGCAGGACAAGCTTCAAGA and CCCAACCCAAAAAGAGTGTGACA (primer pair efficiency: 98%); ACTIN (ACT2) was used as normalizer (23).
Quantification of SUL Silencing.
To quantify SUL silencing spread, pictures of leaves of the corresponding transgenic plants were transformed into black and white images (32 bit), and the area and black/white pixel ratio was calculated using ImageJ.
sRNA Sequencing.
Total RNA extraction was isolated using TRIzol reagent (Invitrogen). Then, total RNAs with an RIN (RNA Integrity Number) value above eight were separated on a 15% TBE-Urea Polyacrylamide Gel (Invitrogen). Small RNAs that are 14–30 nt in length were recovered using ZR small-RNA PAGE Recovery Kit (ZYMO Research). Small-RNA libraries were prepared using NEB Next Multiplex Small RNA Library Prep Set (New England Biolabs). Purified small RNAs were ligated to the 3′ adaptor at 25 °C for 1 h, followed by hybridization with RT Primer at 75 °C for 5 min. The ligation product was then used for 5′ adaptor ligation at 25 °C for 1 h, product of which was used as a template for reverse transcription at 50 °C for 1 h. The cDNA was then amplified using primers that contain indexes. PCR products were purified using MinElute PCR Purification Kit (Qiagen) and 1.6× AMPure XP Beads. Sequencing was performed on a HiSeq2500 (Illumina) according to the manufacturer’s instructions at Genomics Core Facility of Shanghai Center for Plant Stress Biology.
Small-RNA data analyses were performed using a pipeline described in ref. 24. Briefly, raw reads were trimmed using trim_galore v0.4.0 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) to remove the adapter sequences and bases that have a quality score lower than 10. Reads that could not be aligned to structural RNA sequences (rRNA, tRNA, snoRNA, snRNA, etc.) were aligned to the TAIR10 genome using Burrows–Wheeler aligner (25) by allowing one mismatch per read. The number of reads that are mapped to the SUL transgene sequence were summarized and normalized to the structural RNA-removed library size (reads per 10 million).
Supplementary Material
Acknowledgments
We thank Tsuyoshi Nakagawa, Zachary Nimchuk, Yuanzheng Wang, and Alberto Macho for kindly sharing materials; the Proteomics facility at the Shanghai Center for Plant Stress Biology; Wenjie Zeng, Xinyu Jian, Aurora Luque, and Yujing (Ada) Liu for technical assistance; Sebastian Wolf and Alberto Macho for critical reading of the manuscript; and Rafael Jorge Leόn Morcillo and all members in R.L.-D.’s and Alberto Macho’s groups for stimulating discussions and helpful suggestions. This research was supported by the Centre of Excellence for Plant and Microbial Sciences (CEPAMS), established between the John Innes Centre and the Chinese Academy of Sciences and funded by the UK Biotechnology and Biological Sciences Research Council and the Chinese Academy of Sciences, and a Mianshang grant from the National Natural Science Foundation of China (NSFC) (to R.L.-D.) (Grant 31671994). Research in R.L.-D.’s laboratory is funded by the Shanghai Center for Plant Stress Biology of the Chinese Academy of Sciences and the 100 Talent program of the Chinese Academy of Sciences. T.R.-D. is the recipient of a President’s International Fellowship Initiative (PIFI) postdoctoral fellowship (No. 2016PB042) from the Chinese Academy of Sciences. T.J.-G. is sponsored by a CAS-TWAS President’s Fellowship for International PhD students.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715556115/-/DCSupplemental.
References
- 1.Borges F, Martienssen RA. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol. 2015;16:727–741. doi: 10.1038/nrm4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat Genet. 2007;39:848–856. doi: 10.1038/ng2081. [DOI] [PubMed] [Google Scholar]
- 4.Smith LM, et al. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Csorba T, Kontra L, Burgyán J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology. 2015;479–480:85–103. doi: 10.1016/j.virol.2015.02.028. [DOI] [PubMed] [Google Scholar]
- 6.Luna AP, Morilla G, Voinnet O, Bejarano ER. Functional analysis of gene-silencing suppressors from tomato yellow leaf curl disease viruses. Mol Plant Microbe Interact. 2012;25:1294–1306. doi: 10.1094/MPMI-04-12-0094-R. [DOI] [PubMed] [Google Scholar]
- 7.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z, et al. A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. 2016;35:1519–1533. doi: 10.1007/s00299-015-1900-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeYoung BJ, et al. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 2006;45:1–16. doi: 10.1111/j.1365-313X.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu N, et al. BAM 1 and RECEPTOR-LIKE PROTEIN KINASE 2 constitute a signaling pathway and modulate CLE peptide-triggered growth inhibition in Arabidopsis root. New Phytol. 2015;208:1104–1113. doi: 10.1111/nph.13520. [DOI] [PubMed] [Google Scholar]
- 11.Hord CL, Chen C, Deyoung BJ, Clark SE, Ma H. The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell. 2006;18:1667–1680. doi: 10.1105/tpc.105.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soyars CL, James SR, Nimchuk ZL. Ready, aim, shoot: Stem cell regulation of the shoot apical meristem. Curr Opin Plant Biol. 2016;29:163–168. doi: 10.1016/j.pbi.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Carlsbecker A, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nogueira FT, Madi S, Chitwood DH, Juarez MT, Timmermans MC. Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 2007;21:750–755. doi: 10.1101/gad.1528607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chitwood DH, et al. Pattern formation via small RNA mobility. Genes Dev. 2009;23:549–554. doi: 10.1101/gad.1770009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwab R, et al. Endogenous TasiRNAs mediate non-cell autonomous effects on gene regulation in Arabidopsis thaliana. PLoS One. 2009;4:e5980. doi: 10.1371/journal.pone.0005980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lozano-Durán R, et al. The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. Elife. 2013;2:e00983. doi: 10.7554/eLife.00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Q, et al. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: Components and anchoring nucleoporin. Plant J. 2010;61:259–270. doi: 10.1111/j.1365-313X.2009.04048.x. [DOI] [PubMed] [Google Scholar]
- 19.Nakagawa T, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa T, et al. Improved gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem. 2007;71:2095–2100. doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- 21.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 22.Kubo M, et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love AJ, Yun BW, Laval V, Loake GJ, Milner JJ. Cauliflower mosaic virus, a compatible pathogen of Arabidopsis, engages three distinct defense-signaling pathways and activates rapid systemic generation of reactive oxygen species. Plant Physiol. 2005;139:935–948. doi: 10.1104/pp.105.066803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, et al. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc Natl Acad Sci USA. 2013;110:8290–8295. doi: 10.1073/pnas.1300585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.