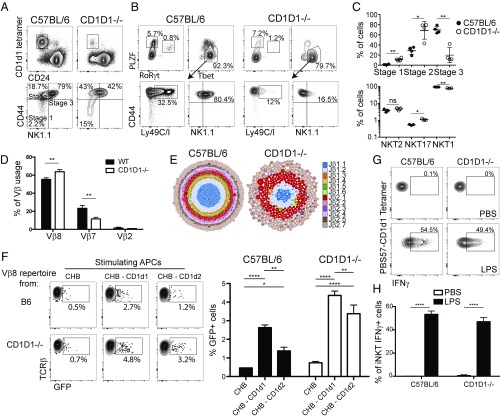
Fig. 3.
Development and repertoire of CD1d2-selected iNKT cells. PBS57-CD1d1 tetramer-reactive thymocytes were enriched from C57BL/6 and pooled CD1D1−/− thymi (≥5 mice per sample) by using MACS beads. Enriched cells were stained with indicated markers and assessed by flow cytometry to characterize specific stages of iNKT-cell development (A) (data representative of n ≥ 3 per group) or functional subsets of iNKT cells as defined by the expression of the transcription factors PLZF, RoRγt, and Tbet (B). (C) Graphs depict mean ± SEM of A and B (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ns, not significant). (D) PBS57-CD1d1 tetramer-reactive thymocytes were enriched by MACS beads from individual B6 or pooled CD1D1−/− thymi (≥5 mice per sample). Enriched cells were stained with PBS57-CD1d1 tetramers, TCRβ, Vβ8, Vβ7, and Vβ2 mAbs and assessed by flow cytometry to characterize Vβ usage (data representative of n ≥ 3 samples per group). Bars depict mean ± SEM of percentage of iNKT cells expressing each Vβ (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ns, not significant). (E) Visual representation of Vβ8 CDR3 sequences from sorted iNKT cells of B6 (Left) and CD1D1−/− (Right) mice. Each CDR3 sequence is represented by a dot, with the size of the dot proportional to the number of times this CDR3 sequence was found with the sample, and each Jβ is represented by a color. Vβ8 rearrangements were amplified by PCR with a V-specific primer and a C-specific reverse primer followed by high-throughput sequencing using the Ion Torrent platform. Sequence analysis was performed with in-house software, and gene identity was assigned on the basis of sequence alignment with published sequences (International ImMunoGeneTics Information System). (F) Vβ8 rearrangements from B6 and CD1D1−/− iNKT cells were amplified and cloned into retroviral plasmids. Retroviruses were produced and used to transduce Vα14-expressing hybridoma. Hybridoma were sorted for similar TCR expression and transduced with NFAT-GFP reporting construct. Antigen presentation assay using the B cell lymphoma CHB cells transfected or not with CD1D1- or CD1D2-expressing constructs was performed. The percentage of GFP+ hybridoma cells from triplicate cultures was recorded by using flow cytometry after 18 h of cultures and quantified (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 by two-way ANOVA). Results are representative of three independent experiments. (G) Intracellular IFN-γ expression of liver iNKT cells 7 h after LPS injection. iNKT cells were identified by using PBS57-CD1d1 tetramer and TCRβ mAbs and stained intracellularly for IFN-γ. Plots are representative of at least three mice per group. (H) Bars depict mean ± SEM of percentage of responding iNKT cells within each strain or condition (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ns, not significant).