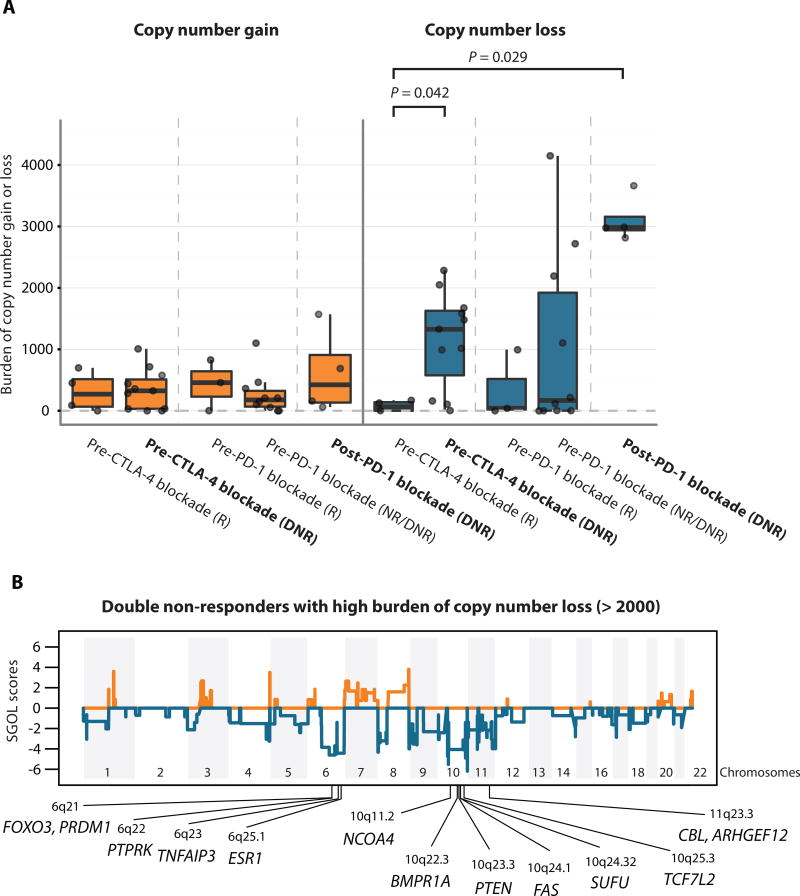
Fig. 2. Copy number loss as a potential resistance mechanism.
(A) Boxplots summarize burden of copy number gain or loss in five groups of interest: responders to CTLA-4 blockade at pre-treatment, pre-CTLA-4 blockade double non-responders, responders to PD-1 blockade at pre-treatment, pre-PD-1 blockade double non-responders, and post-PD-1 blockade double non-responders; median values (lines) and interquartile range (whiskers) are indicated. P values were calculated using a two-sided Mann-Whitney U test. (P = 0.042 for pre-CTLA-4 blockade responders vs. double non-responders, P = 0.029 for pre-CTLA-4 blockade responders vs. post-PD-1 blockade double non-responders, and P > 0.05 for all others) DNR: double non-responders, NR: non-responders, R: responders. In bold are highlighted the Pre-CTLA-4 blockade and Post-PD-1 blockade double non-responder (DNR) groups. (B) Segment Gain or Loss (SGOL) scores were calculated for each copy number segments as sum of log2 copy ratios (tumor/normal) of each copy number segment across all double non-responder samples with burden of copy number loss higher than 2,000 (n=9). Higher positive SGOL scores indicate higher copy number gain of copy number segments and lower negative SGOL scores indicate higher copy number loss of copy number segments. Tumor suppressor genes with recurrent copy number loss are indicated in chromosome 6q, 10q, and 11q23.3.