Introduction
Selenium (Se) was discovered by J.J. Berzelius in 1817 and, while initially identified as a toxin when ingested in large amounts, it was determined to be an essential micronutrient in the 1950s (Boyd, 2011). Since then, Se has been determined to influence human health, and inadequate levels of Se can lead to deleterious effects. Indeed, Se deficiency has been identified as the underlying etiological precipitant of the cardiomyopathy known as Keshan disease, and is also thought to be a key contributor to development of the deforming osteochondropathy, Kashin-Beck disease (Chen, 2012; Yao et al., 2011). Low serum Se levels have also been associated with epilepsy, age-associated neurological disorders, and decreased survival following HIV infection, although a causal role for Se in these diseases has not been proven (Ashrafi et al., 2007; Baum et al., 1997; Zhang et al., 2010).
Once ingested, Se can be present as low molecular weight selenocompounds, or by incorporation into proteins as modified amino acids. One such amino acid modification is selenomethionine (SeMet), which is the primary chemical form of dietary selenium (Burk and Hill, 2015). Once ingested, selenomethionine enters the body’s methionine pool and is incorporated randomly into proteins in place of methionine in proportion to its concentration relative to methionine. However, SeMet can also undergo metabolism in the liver (and other tissues) to yield the selenium-modified cysteine, selenocysteine (Sec), which is degraded by selenocysteine lyase, yielding selenide. The selenide is then metabolized by selenophosphate synthetase to yield selenophosphate, the selenium of which is inserted into serine by selenocysteine synthase to yield selenocysteine attached to the specialized tRNA. The selenocystyl-tRNA then serves to insert the selenocysteine into the nascent protein at the UGA. Collectively, selenocysteine-containing proteins are referred to as selenoproteins (SePs), and are believed to be the primary functional effectors of Se’s effect on human homeostasis and disease. To-date, 25 SePs have been identified in the human genome and 24 in the mouse, including those in the glutathione peroxidase (GPx), idothryonine deiodinase (DIO), and thioredoxin reductase (TrxR) protein families (Gladyshev et al., 2016).
SePs are well-known for their antioxidant roles, as many catalyze oxidation-reduction reactions using the Sec as an active site. The Sec makes them particularly effective enzymes; the Sec selenol group is more fully ionized than the thiol of cysteine (Cys) at physiological pH and has a lower pKa (~5.2) and reduction potential than Cys (Stadtman, 2000). Altogether, these render Sec more reactive than the Cys residue which comprises the active site of many non-selenoprotein enzymes (Stadtman, 1996). Thus, it is perhaps not surprising that the contributions of Se and SePs to human health are most often attributed to their antioxidant functions.
Altered redox homeostasis is believed to be a contributing factor to a number of diseases, and in particular to the development of cancer, as oxidative insult can lead to genomic instability, DNA mutation, tumor initiation, and progression. As Se may reduce oxidative stress by the antioxidant activities of its associated SePs, it was hypothesized that Se supplementation would confer protection against tumor initiation and progression. Indeed, animal tumor models have demonstrated that Se supplementation can reduce the incidence and severity of liver, esophageal, pancreatic, prostatic, colon, and mammary carcinogenesis (Baines et al., 2000; Daoud and Griffin, 1980; Ip et al., 2000; van Rensburg et al., 1986; Wang et al., 2009a; Woutersen et al., 1999). Unfortunately, large clinical trials in human subjects have yielded mixed results; some suggest Se supplementation and/or higher Se status may reduce cancer risk (Clark et al., 1998; Duffield-Lillico et al., 2003; Jacobs et al., 2004) while others fail to correlate serum Se levels with cancer development (Klein et al., 2011; Lippman et al., 2009; Papaioannou et al., 2011; Wallace et al., 2003). Of note, a large international case-control study testing the efficacy of Se and vitamin E supplementation in cancer (SELECT) did not demonstrate a protective effect of Se supplementation on risk of prostate cancer (Klein, et al., 2011; Lippman, et al., 2009), and trial meta-analysis determined that oral administration of Se was not effective in preventing colorectal neoplasia (Lance et al., 2017; Papaioannou, et al., 2011). Furthermore, while not statistically significant, Se supplementation was associated with increased incidence of type-2 diabetes. Thus, the impact of Se supplementation on cancer is now recognized to be a complex issue and careful consideration should be given to untoward “off-target” effects.
Interestingly, one potential explanation for the inconsistent effects of Se supplementation in the above studies is the action of the SePs themselves. Indeed, while SePs have long been thought to protect against cancer development and progression by antioxidant and anti-inflammatory roles, other signaling roles for SePs are becoming increasingly recognized. These other activities, such as modulation of angiogenesis, growth factor signaling, and apoptosis inhibition, may contribute to tumorigenesis in both positive and negative fashions. In this review, we will discuss what is known about the role of a subset of individual SePs in tumorigenesis, with a focus on how these proteins contribute to both pro- and anti-tumorigenic pathways in order to better understand the full effect of Se supplementation on cancer development and growth.
Selenoprotein Production
Expression of SePs is tightly controlled by the SeP translational process, which is highly specialized and with dedicated machinery. Sites of Sec incorporation are identified by the opal codon (UGA) and require the presence of a cis-acting stem-loop sequence in the selenoprotein mRNA 3′ UTR known as a selenocysteine insertion sequence, or SECIS (Walczak et al., 1996). This SECIS site is responsible for recruiting a complex of specialized trans-acting factors dedicated to Sec incorporation, including the Sec-specific tRNA (Sec tRNA) and its required elongation factors (Driscoll and Copeland, 2003; Lee et al., 1990; Low et al., 2000). It is important to note that the SeP translation process is highly dependent on the presence of Se, as the Sec-charged tRNA is only produced upon Se availability (Hatfield et al., 1991). In times of Se deficiency, limited amounts of Sec tRNA results in the UGA codon being read as a premature stop codon, which recruits nonsense mediated decay factors and induces mRNA turnover (Howard et al., 2013; Seyedali and Berry, 2014). More recently, it has been shown that the Sec-specific tRNA can bind cysteine when Se is deficient, resulting in the incorporation of cysteine at UGAs intended to designate Sec (Xu et al., 2010). Whether this process is significant is unknown.
However, in times of selenium deficiency, SeP production is not universally decreased, but rather a “hierarchy” of selenoprotein expression is maintained (Barnes et al., 2009; Schriever et al., 2009). This is due in part to differential SeP mRNA stability based on resistance to nonsense-mediated decay, and SePs responsible for house-keeping functions, such as glutathione peroxidase 4 (GPx4), generally having more stable mRNA than SePs induced in response to stress and injury, such as glutathione peroxidase 1 (GPx1) (Hill et al., 1992; Seyedali and Berry, 2014). Selenoprotein synthesis is also modulated by differential expression of two Sec tRNA isoforms, which are distinguished by the presence or absence of 2′-O-methylribose at position 34. Mice maintained on a normal Se diet have been observed to have increased percentages of mcm5Um-containing Sec tRNA (Um34) as compared to those on Se-deficient chow, suggesting that the Um34 modification is dependent on Se availability (Diamond et al., 1993). Functionally, the two Sec tRNA isoforms are differentially associated with production of specific classes of SePs. The housekeeping SePs essential for survival, such as GPx4, are not dependent on the Um34 modification which allows higher expression in the context of decreased Se availability. Conversely, the stress-induced SePs such as GPx1 and glutathione peroxidase 3 (GPx3) are more responsive to Um34 Sec tRNA and thereby more responsive to Se levels (Hatfield et al., 2006). Indeed, a mutation of the Sec tRNA gene (TRSP) which interferes with the Um34 modification has recently been described in a human subject, where authors note decreased expression of stress-related SePs, while expression of housekeeping SePs is largely preserved (Schoenmakers et al., 2016).
Investigating selenoproteins through SEC tRNA mutations
To determine whether the bulk of Se’s impact was due to low molecular weight selenocompounds, incorporation as selenomethionine, or as incorporation as selenocysteine, early studies sought to specifically investigate the role of the SeP protein family by modifying expression of Sec tRNA (Trsp), thereby globally interfering with selenoprotein biosynthesis. The first such Sec tRNA mouse models [(i6A−) tRNA[Ser]Sec] relied on transgenic expression of mutated Sec tRNA which specifically inhibits Um34 Sec tRNA through a dominant-negative effect (Moustafa et al., 2001). While physiologically normal, mice with global transgene expression were determined to have decreased levels of stress-related SePs, such as GPx1 (Moustafa, et al., 2001). Interestingly, studies using this mouse model were the first to show that SeP expression could directly modify the development of tumorigenesis. In the gut, Sec tRNA transgenic mice were observed to have increased numbers of aberrant crypt foci (ACF), a type of pre-neoplastic colonic lesion, after exposure to the mutagenic agent azoxymethane (AOM) (Irons et al., 2006). Interbreeding of Sec tRNA transgenic mice with a model of prostate cancer (C3(1)/Tag) resulted in accelerated development of prostatic intraepithelial neoplasias (PIN), as well as more aggressive high-grade lesions (Diwadkar-Navsariwala et al., 2006). However, these results were not universal, as Sec tRNA transgenic mice had no change in hepatocellular tumor number or size as compared to WT mice when crossed onto a tumor-prone TGF-α transgenic background (Moustafa et al., 2013). Thus, the role of stress-related SePs in tumorigenesis appears to be dependent on organ and cancer type. Furthermore, these results may also point to differential SeP function in the context of specific oncogenic drivers or undetermined “environmental” co-factors, such as inflammation or microbiota.
The Sec tRNA transgenic model only interfered with translation of a subset of SePs and did not globally inhibit SeP production. However, global knockout (KO) of the mammalian Sec tRNA gene (Trsp) is embryonically lethal, thus a conditional approach has been used to analyze the effects of complete SeP loss (Bosl et al., 1997). Interestingly, even restricting Trsp KO to certain tissues often yielded severe effects, and SeP expression in many tissues, such as the endothelium, cardiac muscle, liver, and skin is apparently required for survival (Carlson et al., 2005; Sengupta et al., 2010; Shrimali et al., 2007). Nevertheless, this model has allowed assessment of SeP contribution to tissue homeostasis as well as a number of malignancies. In the mammary gland, homozygous Trsp loss was associated with increased tumor incidence and shorter survival time after treatment with the carcinogen, 7, 12-dimethylbenzyl[a]antracene (DMBA) (Hudson et al., 2012). Interestingly, while Trsp loss was not sufficient to induce tumors in non-DMBA treated mice, it altered expression of BRCA and p53 in the mammary epithelium and suggests SePs can affect oncogenic pathways in premalignant tissue (Kumaraswamy et al., 2003). In the prostate, Trsp loss alone was sufficient to induce tumorigenesis even in the absence of additional oncogenic alteration (Luchman et al., 2014). These mice presented with widespread PIN lesions by 6 weeks of age, which soon progressed to microinvasive carcinomas characterized by increased proliferation and activation of the mitogen-activated protein kinase (MAPK) pathway. While mice do not live long enough to assess susceptibility to cancer development, Trsp KO in the skin also led to development of widespread proliferation and hyperplasia, which are considered early events in tumor development (Sengupta, et al., 2010). Finally, the Trsp KO model has also been utilized to gain insight into the function of SePs in immune cell population which, in turn, may also influence tumor development. Trsp knockout in myeloid lineages through a LysM-Cre driver led to increased oxidative stress, upregulated transcription of antioxidant enzymes, accumulation of reactive oxygen species, altered expression of extracellular matrix-related genes, diminished migration through matrix, and increased severity in a model of experimental colitis (Carlson, et al., 2005; Kaushal et al., 2014; Suzuki et al., 2008). While the effect of myeloid-specific Trsp loss on tumor development has not been directly analyzed, collectively these results point to a wide-spread tumor-suppressive role for SePs and suggest they inhibit key oncogenic molecular pathways.
Role of Specific Selenoproteins in tumorigenesis
Selenoprotein P
While studies of Se supplementation and global SeP modification suggest SePs broadly suppress tumorigenesis, they do not address the role of specific selenium-containing proteins. Therefore, researchers have generated numerous cell culture and mouse models to investigate individual SePs, such as selenoprotein P (known also as SELENOP, SEPP1, and several other designations (Gladyshev, et al., 2016)). Unlike the majority of SePs which are best characterized by their enzymatic activity, SELENOP is better known for its roles in Se transport. In addition to GPx3, SELENOP is one of the few secreted SePs and is primarily produced in the liver at the main site of Se metabolism (Burk and Hill, 2005). In comparison to most SePs which only have 1 Sec residue, SELENOP is able to incorporate Se in up to 10 Sec residues within its primary structure, with the majority harbored within a Se-rich C-terminal domain. After Sec incorporation, SELENOP is secreted into the plasma and travels to distant tissues where it is taken up via endocytosis and lysosomally degraded to free Se for synthesis of other SePs (Kurokawa et al., 2012; Saito et al., 2004). Indeed, liver-specific Selenop KO revealed a striking 95% reduction in plasma SELENOP levels, which greatly reduced Se levels in distant tissues (Hill et al., 2012). However, it is important to note that SELENOP still retains antioxidant activity through a single N-terminal Sec, which exists within a UXXC motif that catalyzes the oxidation of glutathione (GSH) (Kurokawa et al., 2014b; Saito et al., 1999). Thus, both N- and C-terminal domains contribute to the overall function of SELENOP, making it vital for the production of other selenoproteins within target organs while still serving an antioxidant function.
Because such a large proportion of Se is incorporated into SELENOP, it is frequently measured as a reliable marker for overall Se levels. Thus, a number of epidemiological studies have analyzed SELENOP serum levels and tissue expression to determine their correlation with cancer incidence, aggressiveness, and survival. On the whole, these studies have determined SELENOP to be decreased in various tumor types, including hepatocellular carcinomas, gallbladder and biliary tract cancers, gastric adenocarcinomas, colorectal cancer, and prostate cancer (Al-Taie et al., 2004; Calvo et al., 2002; Gonzalez-Moreno et al., 2011; Hughes et al., 2016; Murawaki et al., 2008; Wang et al., 2009b). Indeed, SELENOP transcript levels are reduced as early as the pre-malignant adenoma stage in the colon (Barrett et al., 2015; Smith et al., 2010). SELENOP expression was also inversely correlated with disease stage in renal, prostate, and colon cancer patients (Meyer et al., 2012; Murawaki, et al., 2008; Outzen et al., 2016). Interestingly, several single nucleotide polymorphisms (SNPs) have also been identified in SELENOP that may attenuate its expression or function, and these SNPs have been associated with cancer risk in the colon, breast, and prostate (Al-Taie et al., 2002; Meplan et al., 2013; Meplan et al., 2010; Pellatt et al., 2013; Peters et al., 2008; Steinbrecher et al., 2010). However, SELENOP expression is not universally decreased in all tumor types. It has been reported to be increased in metastatic melanoma samples as compared to normal melanocytes, and its upregulation has also been associated with poorly-differentiated prostate cancer which is more likely to metastasize (Hassona et al., 2013; Riker et al., 2008).
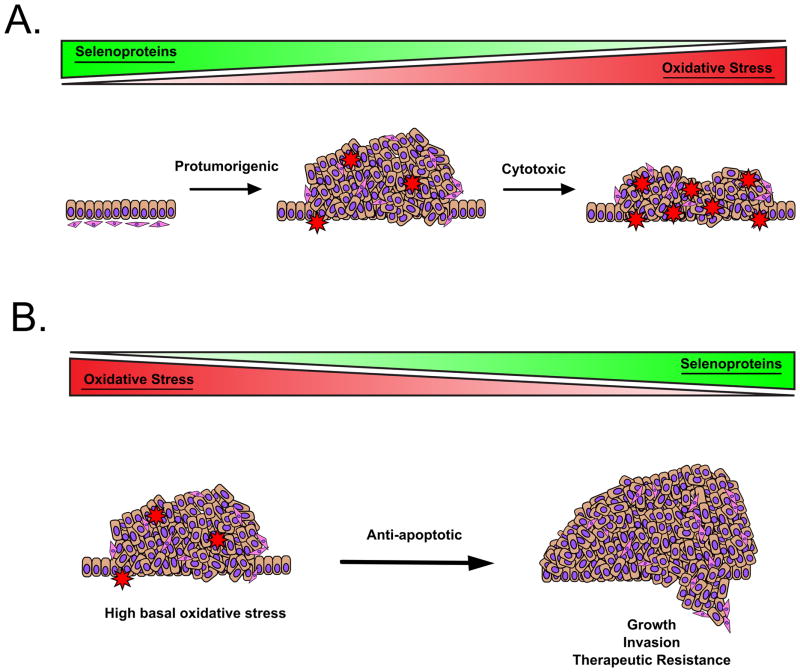
Despite a wealth of expression and epidemiological data, very few studies have directly analyzed SELENOP’s contribution to tumor formation and progression. Using an siRNA approach, Gonzalez-Moreno et al. determined that reductions in SELENOP increased reactive oxygen species (ROS) in prostate cancer cells (Gonzalez-Moreno, et al., 2011). In line with the expression studies, this data supports a role in tumor-suppression by decreasing ROS-mediated mutation and tumor initiation. However, this increase in ROS also rendered cells more susceptible to growth inhibition upon stimulation with hydrogen peroxide (H2O2). Thus, this illustrates a facet of the “double-edged sword” effect of oxidative stress which is highly relevant to studies of SePs in cancer progression; increased oxidative stress can enhance tumorigenesis through genomic instability and mutation, but excess can be toxic and induce tumor cell death (Figure 1A) (Perez et al., 2017). This dichotomy is also highlighted in studies by Barrett et al., in which Selenop KO mice were placed on an azoxymethane/dextran sodium sulfate (AOM/DSS) protocol to induce inflammatory carcinogenesis in the colon (Barrett, et al., 2015). Here, heterozygous Selenop loss augmented tumor number, size, and proliferation, while Selenop null mice were relatively protected from tumorigenesis and displayed smaller tumors with elevated DNA damage and increased apoptosis.
Figure 1.
Due to its roles in Se transport, SELENOP is crucial for formation of other SePs, which may further affect net antioxidant function. Therefore, it can be difficult to determine whether the protumorigenic effects observed with SELENOP reduction are truly specific to SELENOP, or instead driven by loss of other selenium-containing proteins. This question was addressed by utilizing two additional mutant mouse models of SELENOP which express modified genes deficient in either the transport or antioxidant functions (Hill et al., 2007; Kurokawa, et al., 2014b). These studies observed increased tumor numbers with loss of either domain, similar to the results with the heterozygous mice. Interestingly, not only do these results suggest that both antioxidant and transport capabilities functionally contribute to tumorigenesis, but suggest that it is their combined loss which increases oxidative stress to the point of cytotoxicity. Thus, it seems that moderate, but not complete, loss of SELENOP and antioxidant capability is the most detrimental, which further highlights a potential bi-modal effect of SePs and oxidative stress.
While much of SELENOP’s effect on tumor development has been attributed to increased oxidative DNA damage, it can also mediate microenvironmental changes which can further influence tumor cell growth. For example, SELENOP has been found to modify cytokine production, and SELENOP knockdown in the HepG2 hepatoma cell line induced expression of the pro-inflammatory cytokine TNF-α (Yi et al., 2003). Overexpression of SELENOP attenuates TGF-β-mediated transition of fibroblasts to myofibroblasts and reduces their secretion of HGF, VEGF, and IL-6, which can induce tumor cell invasion (Cat et al., 2006). Interestingly, local SELENOP expression is regulated by inflammatory cytokines, as TGF-β, IL-1β, TNF-α, and IFN-γ inhibit SELENOP transcription, while IL-10 induces expression (Bosschaerts et al., 2008; Mostert et al., 1999; Speckmann et al., 2010). SELENOP can also contribute to a protumorigenic microenvironment through altering macrophage polarization and function. Gene expression analysis has determined that SELENOP is one of the most upregulated genes in the M2 macrophage phenotype, which are widely believed to be more pro-tumorigenic than their classically activated M1 counterparts (Bosschaerts, et al., 2008; Solinas et al., 2010). Indeed, macrophages from Selenop heterozygous mice show altered macrophage polarization both in vivo and in vitro and show enhanced macrophage migration in tumors from AOM/DSS treated mice (Barrett, et al., 2015).
In order to take up extracellular SELENOP, cells rely on two known receptors which are differentially expressed based on tissue type. In tissues such as the brain and testes, SELENOP is taken up by apoER2-mediated endocytosis, although in other tissues such as the kidney, the primary SELENOP receptor is megalin. Both receptors are members of the lipoprotein receptor (LRP) family (Burk et al., 2007; Hill, et al., 2007; Olson et al., 2007). However, it is important to note that these receptors are not specific to SELENOP, and each has additional functions that may impact tumor growth. ApoER2, for example, is known to regulate cell motility through interactions with Reelin and Cdc42 (Leemhuis and Bock, 2011), and its knockdown inhibits cell migration in pancreatic cancer cells (Sato et al., 2006). Likewise, megalin is increased in some cancer types such as cerebral lymphomas and melanomas, and in these cases its loss decreases tumor cell proliferation and survival (Andersen et al., 2015; Pedersen et al., 2010). On the other hand, megalin also mediates uptake of Vitamin D (which has been associated with a reduced cancer risk), chemotherapeutic drugs, and drug-sensitizing agents (Christensen and Birn, 2002; Christensen and Willnow, 1999; Deeb et al., 2007). Thus, it is possible that SELENOP may further modulate tumorigenic signaling pathways downstream of these receptors by competitive binding or modifying receptor internalization. Another compelling question is whether SELENOP can bind to other members of the LRP family, as the β-propeller region bound by SELENOP is found on all LRP family members (Kurokawa et al., 2014a). This is especially intriguing in the case of LRP5/6, which functions as the primary receptor for the morphogen WNT3a which elicits numerous pro-tumorigenic growth responses. Interestingly, Se deficiency induces WNT pathway activation, and reduced Selenop enhanced expression of WNT targets and stem cell phenotypes in intestinal tumors and intestinal organoid models, respectively (Barrett, et al., 2015; Kipp et al., 2009). However, whether these effects are mediated by SELENOP interaction with LRP family receptors has yet to be addressed.
Glutathione Peroxidases
The glutathione peroxidase family includes eight members, GPx1-8, and constitute some of the SePs most thoroughly studied for their contributions to tumorigenesis to-date. These proteins catalyze hydroperoxide reduction by using GSH as a reductant, and their antioxidant capabilities are well established. However, not all GPx family members contain the selenocysteine residue required for classification as a SeP. In humans, the selenium-containing GPx family members consist of GPx1, GPx2, GPx3, GPx4, and GPx6, while GPx5, GPx7, and GPx8 contain a Cys in place of Sec (Brigelius-Flohe and Maiorino, 2013). While a SeP in humans, murine GPx6 does not contain a Sec, and its restriction to the adult olfactory system renders its contribution to tumorigenesis unlikely (Dear et al., 1991; Kryukov et al., 2003). Therefore, for this review we will focus on GPx1-4, although the role of GPx family proteins in cancer risk and progression are covered in more depth in an accompanying chapter in this serial.
Of the GPx family members, GPx1 is the most abundant and is widely expressed in the cytosol and mitochondria of various tissue types (Lubos et al., 2011). While all GPx’s catalyze peroxide reduction, GPx1 serves as the family prototype as it contains the full complement of residues for optimal GSH binding (Flohe et al., 2011). As one of the lowest SePs in the “hierarchy of selenoproteins,” GPx1 is highly responsive to Se levels and its expression decreases dramatically in the case of Se deficiency or Sec tRNA modification (Barnes, et al., 2009; Moustafa, et al., 2001). Thus, GPx1 may be one of the SePs most easily modified by Se supplementation. Indeed, GPx1 expression is decreased in many tumor types, and multiple studies have suggested a protective role for GPx1 (Lubos, et al., 2011). For example, overexpression of GPx1 has been found to reduce growth of cancer cells in vitro and in vivo, inhibit pro-survival signaling through the AKT pathway, and attenuate expression of COX2-derived, pro-inflammatory mediators (Capdevila et al., 1995; Handy et al., 2009; Liu et al., 2004; Nasr et al., 2004). On the other hand, studies using mouse models of skin cancer have found that GPx1 overexpressing mice had higher tumor numbers and tumor growth rate, and GPx1 expression has been linked to resistance to chemotherapy and radiation (Gan et al., 2014; Lu et al., 1997; Yang et al., 2015). Interestingly, it is possible that this disparity can be attributed to a further complication of oxidative stress in tumorigenesis (Figure 1B). Indeed, established tumors often have higher levels of basal oxidative stress, and in this case SeP expression can enhance protection from ROS-based apoptosis. Thus, the cumulative effect of GPx1 expression in cancer is not yet clear.
In contrast to GPx1, glutathione peroxidase 2 (GPx2) shows a more localized expression pattern and is most highly expressed in the proliferative crypt compartment of the intestinal epithelium (Florian et al., 2010). Like GPx1, it too appears to have an anti-inflammatory role and can reduce expression of pro-inflammatory cytokines (Banning et al., 2008a). Furthermore, GPx1 and GPx2 appear to synergize, as mice that lack both GPx1 and GPx2 develop ileocolitis and spontaneous tumorigenesis in the small intestine, despite little phenotype being observed with loss of either GPx individually (Esworthy et al., 2001). Interestingly, tumor development was dependent on inflammation driven by interactions with pathogenic microflora, as germ-free mice remained tumor-free (Chu et al., 2004). Placing GPx2 null mice on an inflammatory carcinogenesis protocol also determined that GPx2 null mice had more severe inflammation as well as higher tumor incidence in the colon (Krehl et al., 2012), overall indicating a protective role for GPx2 in colitis and colitis-associated tumorigenesis. However, the effect of GPx2 in non-inflammatory driven cancers appears to be the opposite. GPx2 is often found to be overexpressed in sporadic (i.e. non-hereditary and non-inflammatory) colon cancers as well as cancers from the esophagus and liver, and it appears to promote carcinogenesis in this context (Florian et al., 2001; Lei et al., 2016; Suzuki et al., 2013). GPx2 null mice had decreased ACF formation after AOM treatment, and its knockdown has been shown to decrease growth of HT-29 colon cancer xenografts, patient-derived colon tumor spheroids, and castration-resistant prostate cancer cells (Banning et al., 2008b; Emmink et al., 2014; Muller et al., 2013; Naiki et al., 2014).
While the roles of GPx1 and GPx2 in tumorigenesis appear to be tissue- or context-specific, perhaps the GPx which most clearly functions as a tumor-suppressor is the extracellular family member, GPx3. This GPx is most highly expressed in the kidneys and secreted extracellularly into the plasma, although expression can also be detected in other tissue types (Avissar et al., 1994; Whitin et al., 2002). In tumor cells, GPx3 is a frequent target of hypermethylation, and its downregulation and/or methylation is associated with poorer prognosis and chemotherapeutic resistance in a wide range of cancer types (Falck et al., 2010; Pelosof et al., 2016; Yu et al., 2007). Functionally, GPx3 overexpression has been reported to decrease anchorage-independent growth, xenograft size, and metastasis of prostate cancer cells (Yu, et al., 2007). Interestingly, these authors also report downregulation of c-Met, a tyrosine kinase receptor which mediates signaling downstream of hepatic growth factor (HGF) signaling. Other studies have also determined that GPx3’s anti-tumorigenic function may be through direct interaction with p53-induced gene 3 (PIG3), a p53 target gene which regulates cell death following DNA damage (Wang et al., 2012). In vivo, GPx3 maintained these tumor suppressive functions in the TRAMP model of prostate cancer and the AOM/DSS inflammatory carcinogenesis model of the colon, and in both cases Gpx3 null mice displayed greater tumor number (Barrett et al., 2013; Chang et al., 2016). It is worth noting, however, that knockdown of GPX3 in an established colon cancer line, CaCo-2, both induced apoptosis (in the context of H2O2) and decreased growth in soft agar, supporting a pro-tumorigenic role for GPx3 in these cells (Barrett, et al., 2013). Interestingly, CaCo-2 cells may be an outlier as they express a mutated form of p53, which may interfere with signaling through PIG3.
GPx4 is unique among SePs due to its localization to cellular membranes where it inhibits lipid peroxidation (Maiorino et al., 1991; Ursini et al., 1982). Because of this function, it is required for cellular homeostasis and its loss cannot be compensated by other members of the GPx family. Indeed, loss of GPx4 in vivo is not tolerated during development or in the adult mouse (Yant et al., 2003; Yoo et al., 2012). While conditional GPx4 knockout mice have been generated, the effects of tissue-specific GPx4 knockout on cancer development have not yet been reported (Sengupta et al., 2013). However, studies in cell lines have determined GPx4 may function in a tumor suppressive role, and overexpression reduces tumor growth in fibrosarcoma and pancreatic cancer cells (Heirman et al., 2006; Liu et al., 2006). On the other hand, recent years have also seen exciting developments on the role of GPx4 in a novel non-apoptotic, iron-based cell death pathway known as ferroptosis. In this context, loss of GPx4 has been shown to render tumor cells more sensitive to ferroptosis-inducing compounds, and thus may be a target of therapeutic intervention (Yang et al., 2014; Zhu et al., 2017). Collectively, these results suggest that loss of GPx4 expression functionally mediates tumor cell growth, although further research is needed to determine the full scope of GPx4’s contributions to tumorigenesis.
Thioredoxin reductases
In mammals, the thioredoxin reductase (TrxR) family consists of 3 family members: thioredoxin reductase 1 (TrxR1), thioredoxin reductase 2 (TrxR2), and thioredoxin reductase 3 (TrxR3). While structurally similar, they are broadly distinguished by their differential expression; TrxR1 is predominantly localizes to the cytosol, TrxR2 to the mitochondria, and TrxR3 to the testis, although TrxR1 may also be secreted and TrxR3 can be lowly expressed in other cell types (Soderberg et al., 2000; Su et al., 2005; Sun et al., 2005). Each TrxR family enzyme contains two redox domains, with the C-terminal position harboring a Sec required for activity (Gasdaska et al., 1995; Gladyshev et al., 1996; Lee et al., 2000). Collectively, this group of selenoproteins is responsible for reduction of thioredoxins (Trx), which are ubiquitous small oxidoreductase enzymes capable of reducing disulfide bonds. The Trx enzymes are present in all known organisms and critical for redox-mediated regulation of a wide range of cellular substrates, including ribonuclease reductase, peroxiredoxins, glucocorticoid receptors, and transcription factors such as AP-1 (Powis et al., 2000). As TrxRs are the only known reducer of thioredoxins, they are required for Trx activity and thus indispensable for redox homeostasis. In light of this housekeeping function, it is perhaps not surprising that the expression of TrxR1 and TrxR2 were not as greatly reduced in the Sec tRNA transgenic mouse model as SePs such as GPx1 or GPx3, although their activity is reduced by Se deficiency (Hill et al., 1997; Moustafa, et al., 2001). TrxRs are also responsive Se supplementation, which increases TrxR protein levels and activity and augments antioxidant defense mechanisms (Berggren et al., 1999; Miller et al., 2001). While TrxR is the only reducer of Trx, it is also important to note that TrxR reduces substrates other than Trx. Interestingly, this includes selenite, selenodiglutathione, and methylseleninate (Bjornstedt et al., 1992; Gromer and Gross, 2002; Kumar et al., 1992). Therefore, in addition to being a selenoprotein, TrxR has a critical function in Se metabolism, supports SeP production, and may globally modify SeP function on tumorigenesis.
As TrxR expression is critical to maintaining redox balance, it was first hypothesized to protect against malignant transformation. Indeed, TrxR has been shown to modify many tumor-suppressive pathways, either alone or as a component of the Trx system. For example, TrxR1 promotes maturation of p53 and is able to reactivate oxidized, inactive forms of protein kinase C and PTEN, both of which typically function as tumor suppressors (Kahlos et al., 2003; Lee et al., 2002; Moos et al., 2003). Furthermore, TrxR family members can activate protein tyrosine phosphatases and thereby inhibit receptor tyrosine kinase-induced growth signals, and in this manner TrxR1 has been shown to inhibit Src tyrosine kinase after activation by H2O2 (Shinozaki et al., 2006). TrxR may also inhibit microtubule assembly and thus abrogate tumor cell migration and invasion (Khan and Luduena, 1991). Finally, the increase in vascular endothelial growth factor (VEGF) observed after Se depletion is also associated with TrxR inhibition (Streicher et al., 2004). Therefore, it appears that TrxR can suppress multiple stages of tumor progression, from initiation to growth, invasion, and metastasis.
Surprisingly, in spite of these many tumor-suppressive functions, expression of TrxR family members are instead increased in a variety of cancers and often correlated with more severe disease (Lincoln et al., 2003). On one hand, this upregulation is likely due to TrxR induction by the higher levels of oxidative stress observed in tumor cells, and so does not rule out an anti-tumorigenic role. Yet studies in cell culture systems widely report that this increase in TrxR indeed contributes to tumor cell survival. For example, in multiple myeloma cells, pharmacological inhibition of TrxR with the gold compound, Auranofin, increased ROS-induced apoptosis, decreased activation of NF-κB, and sensitization to NF-κB based therapies (Raninga et al., 2015; Raninga et al., 2016). This is in part due to TrxR’s function as a potent inhibitor of oxidative-stress induced apoptosis via regulation of apoptosis signal-regulating kinase 1 and other redox-sensitive members of the MAPK pathway (Matsuzawa and Ichijo, 2008; Rundlof and Arner, 2004). However, even in studies which do not report widespread apoptosis, TrxR1 loss is often associated with growth arrest, alterations in morphology, and/or anchorage independent growth (Gan et al., 2005; Yoo et al., 2006). In one such, Yoo et al. report that TrxR1 was still required for tumorigenesis (Yoo, et al., 2006). In these experiments, cells with loss of TrxR1 were much less efficient at forming tumor and metastasis in vivo, and tumors that did form still expressed TrxR1 due to loss of the targeting construct. In breast cancer cells, TrxR1 appears to mediate the epithelial-to-mesenchymal transition (EMT), as loss of TrxR1 pushes cells towards an epithelial phenotype while overexpression enhanced invasion in Boyden chamber assays (Bhatia et al., 2016; Dong et al., 2016). Together, these results offer strong evidence that TrxR’s primarily function is that of an oncogene in malignant cells.
Taken together, the effect of TrxR inhibition on tumor cell survival and aggressiveness is so robust that it is currently considered a candidate for therapeutic targeting. Indeed, a TrxR inhibitor, motexafin gadolinium (proposed tradename Xcytrin), was recently evaluated in phase 2 clinical trials for treatment of brain metastases in lung cancer patients. While preclinical data found success in multiple cancer types, it was ultimately not granted FDA approval as it did not offer a significant survival benefit (Brachman et al., 2015; Richards and Mehta, 2007). Gold compounds, such as Auranofin or Aurothioglucose, are other highly potent and well-characterized inhibitors of TrxR (Kean and Kean, 2008). These compounds are currently approved for treatment of rheumatoid arthritis, although studies have also found them to induce apoptosis in a wide variety of cancer cell lines even with fairly low concentrations (Cox et al., 2008). Interestingly, both motexafin gadolinium and various gold compounds show little to no adverse effects, which is surprising considering the Trx system is required for maintaining homeostasis. Thus, the story of pharmacological inhibition of TrxR in cancer is likely not over, and is discussed more thoroughly in the accompanying chapter by Elias Arnér. It is also worth noting that a number of approved cancer pharmaceuticals may derive activity in part by TrxR inhibition. For example, platinum derivatives, such as cisplatin, and chemotherapeutic alkylating agents have been determined to inhibit TrxR, which likely accounts for some degree of their cytotoxicity (Arner et al., 2001; Wang et al., 2008). Taken together, these results suggest that through TrxR activation, Se supplementation may actually promote tumor cell survival in primary and metastatic cancers.
Selenoprotein F
While the above SePs function primarily as cytoplasmic anti-oxidants, a number of SePs localize to the endoplasmic reticulum (ER) and instead can function as regulators of protein folding. Such Se-containing proteins include selenoprotein F (SELENOF, also known as SEP15), type 2 iodothyronine deiodinase (DIO2), selenoprotein M (SELENOM), selenoprotein N (SELENON), selenoprotein K (SELENOK), selenoprotein T (SELENOT), and selenoprotein S (SELENOS) (Davis et al., 2012). While the role of the majority of these ER-localized SePs in tumorigenesis are still unclear, one, SELENOF, has been reported to mediate cancer cell growth and development.
SELENOF is a 15kDa thioredoxin-like oxidoreductase which interacts with the ER folding sensor UDP-glucose:glycoprotein glucosyltransferase in order to mediate quality control of protein disulfide bonds. While originally described in human T cells, it is also expressed in numerous epithelial tissues such as the liver and prostate (Gladyshev et al., 1998). Interestingly, SELENOF is located on a locus of chromosome 1p31 which is often deleted in human cancers, and SELENOF is downregulated in gastric cancers, mesotheliomas, hepatocellular carcinomas, prostate cancers, and colorectal cancers (Apostolou et al., 2004; Irons et al., 2010; Kumaraswamy et al., 2000; Lan et al., 2016). SELENOF also harbors two polymorphic sites at positions 811 (C/T) and 1125 (G/A), with the latter localized to the SECIS insertion element (Gladyshev, et al., 1998). Thus, the A1125 appears to have lower Sec incorporation efficiency and is less sensitive to Se levels (Apostolou, et al., 2004). This particular allele is associated with increased risk of breast and head and neck cancers in African Americans, although among smokers, those harboring a GG or GA1125 variant were at higher risk for lung cancer than those with AA1125 (Hu et al., 2001; Jablonska et al., 2008). SELENOF is a functional homolog of SELENOM (Reeves and Hoffmann, 2009), which is downregulated in cholangiocarcinoma and low expression is correlated with worse prognosis (Dai et al., 2016). However, SELENOM’s functional contribution to tumorigenesis remains largely unknown.
Despite being one of the most recently identified SePs, the effect of SELENOF expression on tumor growth has been investigated in a number of cancer models. Despite frequent reports of its reduction in tumor cells and tumor cell lines, these studies uniformly report that SELENOF loss attenuates pro-tumorigenic phenotypes. Downregulation of SELENOF in cancer cell lines resulted in upregulation of the cell cycle inhibitors p21 and p27, and lead to decreased proliferation and growth arrest in colon and liver cancer cells (Bang et al., 2015a; Irons, et al., 2010; Tsuji et al., 2011). These studies also determined that SELENOF loss decreased growth in soft-agar and induced aberrant activation of Rho signaling (Bang et al., 2015b; Irons, et al., 2010; Tsuji, et al., 2011). Colon cancer cells with SELENOF knock-down were also less likely to form tumors and had a striking decrease in pulmonary metastases (Irons, et al., 2010). Likewise, Tsuji et. al observed that SELENOF loss prevented the formation of aberrant crypt foci following AOM injection and induced IFN-γ and STAT1-regulated gene expression (Tsuji et al., 2012). Collectively, these results are somewhat counterintuitive, as sustained ER stress and abnormal protein folding are often associated with aggressive features and drug-resistance, and SELENOF KO indeed display phenotypes attributed to abnormal protein folding (Cubillos-Ruiz et al., 2017; Kasaikina et al., 2011). However, these effects on protein folding may be overshadowed by robust activation of the STAT1 pathway, which induces apoptosis and anti-tumoral immunity. It also remains possible that effects of SELENOF are tissue specific, as its knockdown in mesothelioma cells abrogated apoptosis and decreased cell growth induced by treatment with Se, while loss of SELENOF in lung cancer cells had no discernable effects (Apostolou, et al., 2004; Irons, et al., 2010). Thus, additional studies are necessary to determine the full contribution of SELENOF to multiple cancer types.
Considerations and Conclusions
A wealth of data exists correlating levels of Se and/or expression of selenium-containing proteins with cancer risk and tumor aggressiveness. While these data suggest the effects of Se on tumor biology can partially be attributed to SePs, mechanistic studies to determine exactly how these proteins modify tumor growth are still comparatively lacking. In this review, we have discussed what is known about just a fraction of the SeP family, which were chosen due to their more established connections to tumor growth. However, a number of SePs still remain relatively uncharacterized in the context of tumorigenesis, such as members of the iodothyronine deiodinase family (DIO1, DIO2, and DIO3), SELENOK, SELENOS, and SELENOT. Future studies may identify contributions of these proteins to oncogenic pathways, as well. It is also worth noting that Se can function through Se-binding proteins which do not incorporate a selenocysteine. One such protein, selenium-binding protein 1, has been reported to function as a tumor suppressor, and reduced levels are correlated with poor clinical outcome in various cancer types (Yang and Diamond, 2013). SECIS-binding protein 2 is a major component of the selenocysteine insertion machinery, thus its expression can broadly influence SeP production (Fletcher et al., 2001). While these proteins are not classified as SePs, their expression or activity may broadly impact tumorigenesis in a Se-dependent manner.
Overall, tissue-specific and context-specific analysis appears crucial to our understanding of SeP function in cancer progression. Many SePs, such as GPx1 and TrxR1, are widely expressed across cell and tissue types, while SELENOP and GPx3 are secreted into the plasma. Thus, these proteins can affect numerous cell types, although research suggests not all are influenced equally. For example, GPx1 overexpression in a mouse model of skin cancer found increased tumor growth, yet reduced tumor growth was observed in the context of pancreatic cancer. Knockdown of SELENOF induced apoptosis in mesothelioma cells yet had no effect on growth of lung cancer cells. As pathology, aggressiveness, and oncogenic drivers can vary across tissues, and even across molecular subtypes of the same cancer, the analysis of tissue-specific contributions will greatly aid our understanding of SePs’ role in carcinogenesis. Further testing will also need to address disparities in the context of alternative molecular drivers and mutagenic alterations. For example, the broad tumor suppressive abilities of GPx3 were lost in the context of p53 mutation (Barrett, et al., 2013). However, the effect of p53 mutation on the pro-apoptotic function of other SePs remains unclear.
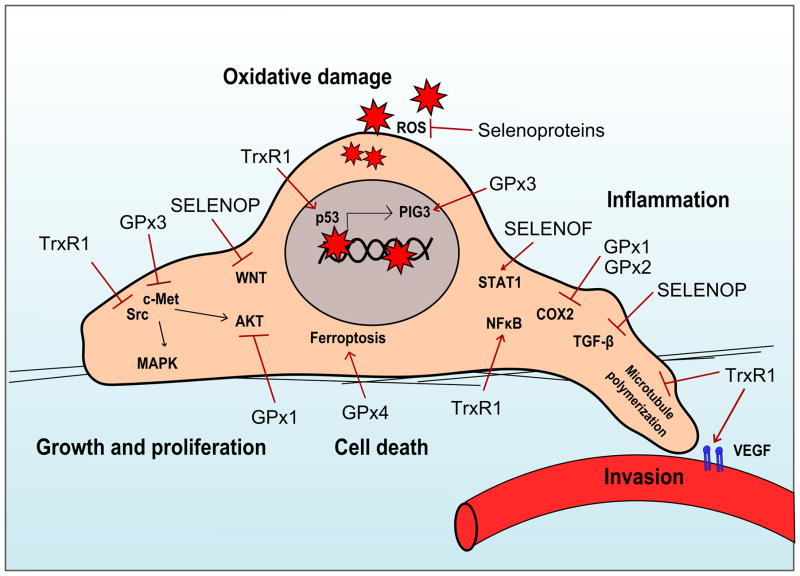
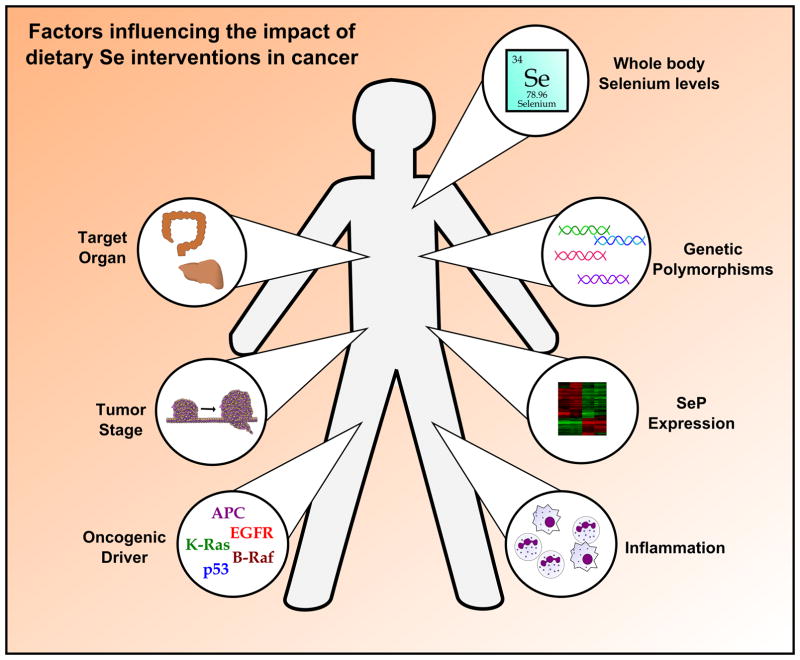
Collectively, these data suggest that SePs are potent modifiers of tumorigenesis, although their action can vary greatly. Numerous epidemiological, in vitro, and in vivo studies are in agreement that SePs can inhibit tumor development by abrogating oxidative insults and mutagenesis, particularly in the context of inflammatory-driven cancers. However, in a tumor cell with higher basal levels of oxidative injury, this SeP activity can render tumor cells resistant to apoptosis and even chemotherapeutic intervention. Furthermore, while linked by the presence of a Sec, SePs are quite varied in their structure and function, and can individually influence numerous oncogenic pathways, such as MAPK, AKT, VEGF, and c-Met (Figure 2). Thus, Se supplementation remains an area of considerable debate, as global modification of selenium-containing proteins may not always be beneficial, and indeed it may be harmful in established tumors. Much work remains to be done to define the precise biologic mechanisms by which individual SePs influence oncogenic programs, taking into account molecular drivers, tissue types, and tumor stage. Once delineated, this knowledge could be overlaid with patient-specific characteristics, such as tissue Se status, SeP levels in target organs, and the presence of genetic polymorphisms in individual SePs (Figure 3). Ultimately, this could determine the benefit of Se supplementation for individual disease prevention and treatment, and while complex, provide enormous benefit for human health.
Figure 2.
Figure 3.
Acknowledgments
The authors gratefully acknowledge Dr. Raymond Burk for his expertise and insightful comments on this manuscript.
References
- Al-Taie OH, Seufert J, Mork H, Treis H, Mentrup B, Thalheimer A, Starostik P, Abel J, Scheurlen M, Kohrle J, Jakob F. Eur J Hum Genet. 2002;10:499–504. doi: 10.1038/sj.ejhg.5200811. [DOI] [PubMed] [Google Scholar]
- Al-Taie OH, Uceyler N, Eubner U, Jakob F, Mork H, Scheurlen M, Brigelius-Flohe R, Schottker K, Abel J, Thalheimer A, Katzenberger T, Illert B, Melcher R, Kohrle J. Nutr Cancer. 2004;48:6–14. doi: 10.1207/s15327914nc4801_2. [DOI] [PubMed] [Google Scholar]
- Andersen RK, Hammer K, Hager H, Christensen JN, Ludvigsen M, Honore B, Thomsen MB, Madsen M. Pigment Cell Melanoma Res. 2015;28:267–80. doi: 10.1111/pcmr.12352. [DOI] [PubMed] [Google Scholar]
- Anestal K, Prast-Nielsen S, Cenas N, Arner ES. PLoS One. 2008;3:e1846. doi: 10.1371/journal.pone.0001846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou S, Klein JO, Mitsuuchi Y, Shetler JN, Poulikakos PI, Jhanwar SC, Kruger WD, Testa JR. Oncogene. 2004;23:5032–40. doi: 10.1038/sj.onc.1207683. [DOI] [PubMed] [Google Scholar]
- Arner ES, Nakamura H, Sasada T, Yodoi J, Holmgren A, Spyrou G. Free Radic Biol Med. 2001;31:1170–8. doi: 10.1016/s0891-5849(01)00698-0. [DOI] [PubMed] [Google Scholar]
- Arner ES, Sarioglu H, Lottspeich F, Holmgren A, Bock A. J Mol Biol. 1999;292:1003–16. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- Ashrafi MR, Shams S, Nouri M, Mohseni M, Shabanian R, Yekaninejad MS, Chegini N, Khodadad A, Safaralizadeh R. Epilepsia. 2007;48:1750–5. doi: 10.1111/j.1528-1167.2007.01143.x. [DOI] [PubMed] [Google Scholar]
- Avissar N, Ornt DB, Yagil Y, Horowitz S, Watkins RH, Kerl EA, Takahashi K, Palmer IS, Cohen HJ. Am J Physiol. 1994;266:C367–75. doi: 10.1152/ajpcell.1994.266.2.C367. [DOI] [PubMed] [Google Scholar]
- Baines AT, Holubec H, Basye JL, Thorne P, Bhattacharyya AK, Spallholz J, Shriver B, Cui H, Roe D, Clark LC, Earnest DL, Nelson MA. Cancer Lett. 2000;160:193–8. doi: 10.1016/s0304-3835(00)00585-1. [DOI] [PubMed] [Google Scholar]
- Bang J, Huh JH, Na JW, Lu Q, Carlson BA, Tobe R, Tsuji PA, Gladyshev VN, Hatfield DL, Lee BJ. Mol Cells. 2015a;38:457–65. doi: 10.14348/molcells.2015.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang J, Jang M, Huh JH, Na JW, Shim M, Carlson BA, Tobe R, Tsuji PA, Gladyshev VN, Hatfield DL, Lee BJ. Biochem Biophys Res Commun. 2015b;456:884–90. doi: 10.1016/j.bbrc.2014.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banning A, Florian S, Deubel S, Thalmann S, Muller-Schmehl K, Jacobasch G, Brigelius-Flohe R. Antioxid Redox Signal. 2008a;10:1491–500. doi: 10.1089/ars.2008.2047. [DOI] [PubMed] [Google Scholar]
- Banning A, Kipp A, Schmitmeier S, Lowinger M, Florian S, Krehl S, Thalmann S, Thierbach R, Steinberg P, Brigelius-Flohe R. Cancer Res. 2008b;68:9746–53. doi: 10.1158/0008-5472.CAN-08-1321. [DOI] [PubMed] [Google Scholar]
- Barnes KM, Evenson JK, Raines AM, Sunde RA. J Nutr. 2009;139:199–206. doi: 10.3945/jn.108.098624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CW, Ning W, Chen X, Smith JJ, Washington MK, Hill KE, Coburn LA, Peek RM, Chaturvedi R, Wilson KT, Burk RF, Williams CS. Cancer Res. 2013;73:1245–55. doi: 10.1158/0008-5472.CAN-12-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CW, Reddy VK, Short SP, Motley AK, Lintel MK, Bradley AM, Freeman T, Vallance J, Ning W, Parang B, Poindexter SV, Fingleton B, Chen X, Washington MK, Wilson KT, Shroyer NF, Hill KE, Burk RF, Williams CS. J Clin Invest. 2015;125:2646–60. doi: 10.1172/JCI76099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum MK, Shor-Posner G, Lai S, Zhang G, Lai H, Fletcher MA, Sauberlich H, Page JB. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;15:370–4. doi: 10.1097/00042560-199708150-00007. [DOI] [PubMed] [Google Scholar]
- Berggren MM, Mangin JF, Gasdaka JR, Powis G. Biochem Pharmacol. 1999;57:187–93. doi: 10.1016/s0006-2952(98)00283-4. [DOI] [PubMed] [Google Scholar]
- Bhatia M, Lovitt CJ, Raninga PV, Avery VM, Di Trapani G, Tonissen KF. Eur J Cell Biol. 2016;95:378–388. doi: 10.1016/j.ejcb.2016.08.003. [DOI] [PubMed] [Google Scholar]
- Bjornstedt M, Kumar S, Holmgren A. J Biol Chem. 1992;267:8030–4. [PubMed] [Google Scholar]
- Bosl MR, Takaku K, Oshima M, Nishimura S, Taketo MM. Proc Natl Acad Sci U S A. 1997;94:5531–4. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosschaerts T, Guilliams M, Noel W, Herin M, Burk RF, Hill KE, Brys L, Raes G, Ghassabeh GH, De Baetselier P, Beschin A. J Immunol. 2008;180:6168–75. doi: 10.4049/jimmunol.180.9.6168. [DOI] [PubMed] [Google Scholar]
- Boyd R. Nat Chem. 2011;3:570. doi: 10.1038/nchem.1076. [DOI] [PubMed] [Google Scholar]
- Brachman DG, Pugh SL, Ashby LS, Thomas TA, Dunbar EM, Narayan S, Robins HI, Bovi JA, Rockhill JK, Won M, Curran WP. Int J Radiat Oncol Biol Phys. 2015;91:961–7. doi: 10.1016/j.ijrobp.2014.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigelius-Flohe R, Maiorino M. Biochim Biophys Acta. 2013;1830:3289–303. doi: 10.1016/j.bbagen.2012.11.020. [DOI] [PubMed] [Google Scholar]
- Burk RF, Hill KE. Annu Rev Nutr. 2005;25:215–35. doi: 10.1146/annurev.nutr.24.012003.132120. [DOI] [PubMed] [Google Scholar]
- Burk RF, Hill KE. Annu Rev Nutr. 2015;35:109–34. doi: 10.1146/annurev-nutr-071714-034250. [DOI] [PubMed] [Google Scholar]
- Burk RF, Hill KE, Olson GE, Weeber EJ, Motley AK, Winfrey VP, Austin LM. J Neurosci. 2007;27:6207–11. doi: 10.1523/JNEUROSCI.1153-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo A, Xiao N, Kang J, Best CJ, Leiva I, Emmert-Buck MR, Jorcyk C, Green JE. Cancer Res. 2002;62:5325–35. [PubMed] [Google Scholar]
- Capdevila JH, Morrow JD, Belosludtsev YY, Beauchamp DR, DuBois RN, Falck JR. Biochemistry. 1995;34:3325–37. doi: 10.1021/bi00010a023. [DOI] [PubMed] [Google Scholar]
- Carlson BA, Xu XM, Gladyshev VN, Hatfield DL. J Biol Chem. 2005;280:5542–8. doi: 10.1074/jbc.M411725200. [DOI] [PubMed] [Google Scholar]
- Cat B, Stuhlmann D, Steinbrenner H, Alili L, Holtkotter O, Sies H, Brenneisen P. J Cell Sci. 2006;119:2727–38. doi: 10.1242/jcs.03011. [DOI] [PubMed] [Google Scholar]
- Chang SN, Lee JM, Oh H, Park JH. Prostate. 2016;76:1387–98. doi: 10.1002/pros.23223. [DOI] [PubMed] [Google Scholar]
- Chen J. Asia Pac J Clin Nutr. 2012;21:320–6. [PubMed] [Google Scholar]
- Christensen EI, Birn H. Nat Rev Mol Cell Biol. 2002;3:256–66. doi: 10.1038/nrm778. [DOI] [PubMed] [Google Scholar]
- Christensen EI, Willnow TE. J Am Soc Nephrol. 1999;10:2224–36. doi: 10.1681/ASN.V10102224. [DOI] [PubMed] [Google Scholar]
- Chu FF, Esworthy RS, Chu PG, Longmate JA, Huycke MM, Wilczynski S, Doroshow JH. Cancer Res. 2004;64:962–8. doi: 10.1158/0008-5472.can-03-2272. [DOI] [PubMed] [Google Scholar]
- Clark LC, Dalkin B, Krongrad A, Combs GF, Jr, Turnbull BW, Slate EH, Witherington R, Herlong JH, Janosko E, Carpenter D, Borosso C, Falk S, Rounder J. Br J Urol. 1998;81:730–4. doi: 10.1046/j.1464-410x.1998.00630.x. [DOI] [PubMed] [Google Scholar]
- Cox AG, Brown KK, Arner ES, Hampton MB. Biochem Pharmacol. 2008;76:1097–109. doi: 10.1016/j.bcp.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Cubillos-Ruiz JR, Bettigole SE, Glimcher LH. Cell. 2017;168:692–706. doi: 10.1016/j.cell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Thongchot S, Dokduang H, Loilome W, Khuntikeo N, Titapun A, Ungarreevittaya P, Yongvanit P, Techasen A, Namwat N. Anticancer Res. 2016;36:5981–5988. doi: 10.21873/anticanres.11186. [DOI] [PubMed] [Google Scholar]
- Daoud AH, Griffin AC. Cancer Lett. 1980;9:299–304. doi: 10.1016/0304-3835(80)90021-x. [DOI] [PubMed] [Google Scholar]
- Davis CD, Tsuji PA, Milner JA. Annu Rev Nutr. 2012;32:73–95. doi: 10.1146/annurev-nutr-071811-150740. [DOI] [PubMed] [Google Scholar]
- Dear TN, Campbell K, Rabbitts TH. Biochemistry. 1991;30:10376–82. doi: 10.1021/bi00107a003. [DOI] [PubMed] [Google Scholar]
- Deeb KK, Trump DL, Johnson CS. Nat Rev Cancer. 2007;7:684–700. doi: 10.1038/nrc2196. [DOI] [PubMed] [Google Scholar]
- Diamond AM, Choi IS, Crain PF, Hashizume T, Pomerantz SC, Cruz R, Steer CJ, Hill KE, Burk RF, McCloskey JA, Hatfield DL. J Biol Chem. 1993;268:14215–23. [PubMed] [Google Scholar]
- Diwadkar-Navsariwala V, Prins GS, Swanson SM, Birch LA, Ray VH, Hedayat S, Lantvit DL, Diamond AM. Proc Natl Acad Sci U S A. 2006;103:8179–84. doi: 10.1073/pnas.0508218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Zhang L, Sun R, Liu J, Yin H, Li X, Zheng X, Zeng H. Sci Rep. 2016;6:36860. doi: 10.1038/srep36860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll DM, Copeland PR. Annu Rev Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- Duffield-Lillico AJ, Dalkin BL, Reid ME, Turnbull BW, Slate EH, Jacobs ET, Marshall JR, Clark LC. BJU Int. 2003;91:608–12. doi: 10.1046/j.1464-410x.2003.04167.x. [DOI] [PubMed] [Google Scholar]
- Emmink BL, Laoukili J, Kipp AP, Koster J, Govaert KM, Fatrai S, Verheem A, Steller EJ, Brigelius-Flohe R, Jimenez CR, Borel Rinkes IH, Kranenburg O. Cancer Res. 2014;74:6717–30. doi: 10.1158/0008-5472.CAN-14-1645. [DOI] [PubMed] [Google Scholar]
- Esworthy RS, Aranda R, Martin MG, Doroshow JH, Binder SW, Chu FF. Am J Physiol Gastrointest Liver Physiol. 2001;281:G848–55. doi: 10.1152/ajpgi.2001.281.3.G848. [DOI] [PubMed] [Google Scholar]
- Falck E, Karlsson S, Carlsson J, Helenius G, Karlsson M, Klinga-Levan K. Cancer Cell Int. 2010;10:46. doi: 10.1186/1475-2867-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JE, Copeland PR, Driscoll DM, Krol A. RNA. 2001;7:1442–53. [PMC free article] [PubMed] [Google Scholar]
- Flohe L, Toppo S, Cozza G, Ursini F. Antioxid Redox Signal. 2011;15:763–80. doi: 10.1089/ars.2010.3397. [DOI] [PubMed] [Google Scholar]
- Florian S, Krehl S, Loewinger M, Kipp A, Banning A, Esworthy S, Chu FF, Brigelius-Flohe R. Free Radic Biol Med. 2010;49:1694–702. doi: 10.1016/j.freeradbiomed.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florian S, Wingler K, Schmehl K, Jacobasch G, Kreuzer OJ, Meyerhof W, Brigelius-Flohe R. Free Radic Res. 2001;35:655–63. doi: 10.1080/10715760100301181. [DOI] [PubMed] [Google Scholar]
- Gan L, Yang XL, Liu Q, Xu HB. J Cell Biochem. 2005;96:653–64. doi: 10.1002/jcb.20585. [DOI] [PubMed] [Google Scholar]
- Gan X, Chen B, Shen Z, Liu Y, Li H, Xie X, Xu X, Li H, Huang Z, Chen J. Int J Clin Exp Med. 2014;7:2530–40. [PMC free article] [PubMed] [Google Scholar]
- Gasdaska PY, Gasdaska JR, Cochran S, Powis G. FEBS Lett. 1995;373:5–9. doi: 10.1016/0014-5793(95)01003-w. [DOI] [PubMed] [Google Scholar]
- Gladyshev VN, Arner ES, Berry MJ, Brigelius-Flohe R, Bruford EA, Burk RF, Carlson BA, Castellano S, Chavatte L, Conrad M, Copeland PR, Diamond AM, Driscoll DM, Ferreiro A, Flohe L, Green FR, Guigo R, Handy DE, Hatfield DL, Hesketh J, Hoffmann PR, Holmgren A, Hondal RJ, Howard MT, Huang K, Kim HY, Kim IY, Kohrle J, Krol A, Kryukov GV, Lee BJ, Lee BC, Lei XG, Liu Q, Lescure A, Lobanov AV, Loscalzo J, Maiorino M, Mariotti M, Sandeep Prabhu K, Rayman MP, Rozovsky S, Salinas G, Schmidt EE, Schomburg L, Schweizer U, Simonovic M, Sunde RA, Tsuji PA, Tweedie S, Ursini F, Whanger PD, Zhang Y. J Biol Chem. 2016;291:24036–24040. doi: 10.1074/jbc.M116.756155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev VN, Jeang KT, Stadtman TC. Proc Natl Acad Sci U S A. 1996;93:6146–51. doi: 10.1073/pnas.93.12.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev VN, Jeang KT, Wootton JC, Hatfield DL. J Biol Chem. 1998;273:8910–5. doi: 10.1074/jbc.273.15.8910. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Moreno O, Boque N, Redrado M, Milagro F, Campion J, Endermann T, Takahashi K, Saito Y, Catena R, Schomburg L, Calvo A. Prostate. 2011;71:824–34. doi: 10.1002/pros.21298. [DOI] [PubMed] [Google Scholar]
- Gromer S, Gross JH. J Biol Chem. 2002;277:9701–6. doi: 10.1074/jbc.M109234200. [DOI] [PubMed] [Google Scholar]
- Handy DE, Lubos E, Yang Y, Galbraith JD, Kelly N, Zhang YY, Leopold JA, Loscalzo J. J Biol Chem. 2009;284:11913–21. doi: 10.1074/jbc.M900392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassona Y, Cirillo N, Lim KP, Herman A, Mellone M, Thomas GJ, Pitiyage GN, Parkinson EK, Prime SS. Carcinogenesis. 2013;34:1286–95. doi: 10.1093/carcin/bgt035. [DOI] [PubMed] [Google Scholar]
- Hatfield D, Lee BJ, Hampton L, Diamond AM. Nucleic Acids Res. 1991;19:939–43. doi: 10.1093/nar/19.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield DL, Carlson BA, Xu XM, Mix H, Gladyshev VN. Prog Nucleic Acid Res Mol Biol. 2006;81:97–142. doi: 10.1016/S0079-6603(06)81003-2. [DOI] [PubMed] [Google Scholar]
- Heirman I, Ginneberge D, Brigelius-Flohe R, Hendrickx N, Agostinis P, Brouckaert P, Rottiers P, Grooten J. Free Radic Biol Med. 2006;40:285–94. doi: 10.1016/j.freeradbiomed.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Hill KE, Lyons PR, Burk RF. Biochem Biophys Res Commun. 1992;185:260–3. doi: 10.1016/s0006-291x(05)80984-2. [DOI] [PubMed] [Google Scholar]
- Hill KE, McCollum GW, Boeglin ME, Burk RF. Biochem Biophys Res Commun. 1997;234:293–5. doi: 10.1006/bbrc.1997.6618. [DOI] [PubMed] [Google Scholar]
- Hill KE, Wu S, Motley AK, Stevenson TD, Winfrey VP, Capecchi MR, Atkins JF, Burk RF. J Biol Chem. 2012;287:40414–24. doi: 10.1074/jbc.M112.421404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KE, Zhou J, Austin LM, Motley AK, Ham AJ, Olson GE, Atkins JF, Gesteland RF, Burk RF. J Biol Chem. 2007;282:10972–80. doi: 10.1074/jbc.M700436200. [DOI] [PubMed] [Google Scholar]
- Howard MT, Carlson BA, Anderson CB, Hatfield DL. J Biol Chem. 2013;288:19401–13. doi: 10.1074/jbc.M113.481051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YJ, Korotkov KV, Mehta R, Hatfield DL, Rotimi CN, Luke A, Prewitt TE, Cooper RS, Stock W, Vokes EE, Dolan ME, Gladyshev VN, Diamond AM. Cancer Res. 2001;61:2307–10. [PubMed] [Google Scholar]
- Hudson TS, Carlson BA, Hoeneroff MJ, Young HA, Sordillo L, Muller WJ, Hatfield DL, Green JE. Carcinogenesis. 2012;33:1225–30. doi: 10.1093/carcin/bgs129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DJ, Duarte-Salles T, Hybsier S, Trichopoulou A, Stepien M, Aleksandrova K, Overvad K, Tjonneland A, Olsen A, Affret A, Fagherazzi G, Boutron-Ruault MC, Katzke V, Kaaks R, Boeing H, Bamia C, Lagiou P, Peppa E, Palli D, Krogh V, Panico S, Tumino R, Sacerdote C, Bueno-de-Mesquita HB, Peeters PH, Engeset D, Weiderpass E, Lasheras C, Agudo A, Sanchez MJ, Navarro C, Ardanaz E, Dorronsoro M, Hemmingsson O, Wareham NJ, Khaw KT, Bradbury KE, Cross AJ, Gunter M, Riboli E, Romieu I, Schomburg L, Jenab M. Am J Clin Nutr. 2016;104:406–14. doi: 10.3945/ajcn.116.131672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip C, Thompson HJ, Ganther HE. Cancer Epidemiol Biomarkers Prev. 2000;9:49–54. [PubMed] [Google Scholar]
- Irons R, Carlson BA, Hatfield DL, Davis CD. J Nutr. 2006;136:1311–7. doi: 10.1093/jn/136.5.1311. [DOI] [PubMed] [Google Scholar]
- Irons R, Tsuji PA, Carlson BA, Ouyang P, Yoo MH, Xu XM, Hatfield DL, Gladyshev VN, Davis CD. Cancer Prev Res (Phila) 2010;3:630–9. doi: 10.1158/1940-6207.CAPR-10-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonska E, Gromadzinska J, Sobala W, Reszka E, Wasowicz W. Eur J Nutr. 2008;47:47–54. doi: 10.1007/s00394-008-0696-9. [DOI] [PubMed] [Google Scholar]
- Jacobs ET, Jiang R, Alberts DS, Greenberg ER, Gunter EW, Karagas MR, Lanza E, Ratnasinghe L, Reid ME, Schatzkin A, Smith-Warner SA, Wallace K, Martinez ME. J Natl Cancer Inst. 2004;96:1669–75. doi: 10.1093/jnci/djh310. [DOI] [PubMed] [Google Scholar]
- Kahlos K, Zhang J, Block ER, Patel JM. Mol Cell Biochem. 2003;254:47–54. doi: 10.1023/a:1027380828645. [DOI] [PubMed] [Google Scholar]
- Kasaikina MV, Fomenko DE, Labunskyy VM, Lachke SA, Qiu W, Moncaster JA, Zhang J, Wojnarowicz MW, Jr, Natarajan SK, Malinouski M, Schweizer U, Tsuji PA, Carlson BA, Maas RL, Lou MF, Goldstein LE, Hatfield DL, Gladyshev VN. J Biol Chem. 2011;286:33203–12. doi: 10.1074/jbc.M111.259218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal N, Kudva AK, Patterson AD, Chiaro C, Kennett MJ, Desai D, Amin S, Carlson BA, Cantorna MT, Prabhu KS. J Immunol. 2014;193:3683–92. doi: 10.4049/jimmunol.1400347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean WF, Kean IR. Inflammopharmacology. 2008;16:112–25. doi: 10.1007/s10787-007-0021-x. [DOI] [PubMed] [Google Scholar]
- Khan IA, Luduena RF. Biochim Biophys Acta. 1991;1076:289–97. doi: 10.1016/0167-4838(91)90280-d. [DOI] [PubMed] [Google Scholar]
- Kipp A, Banning A, van Schothorst EM, Meplan C, Schomburg L, Evelo C, Coort S, Gaj S, Keijer J, Hesketh J, Brigelius-Flohe R. Mol Nutr Food Res. 2009;53:1561–72. doi: 10.1002/mnfr.200900105. [DOI] [PubMed] [Google Scholar]
- Klein EA, Thompson IM, Jr, Tangen CM, Crowley JJ, Lucia MS, Goodman PJ, Minasian LM, Ford LG, Parnes HL, Gaziano JM, Karp DD, Lieber MM, Walther PJ, Klotz L, Parsons JK, Chin JL, Darke AK, Lippman SM, Goodman GE, Meyskens FL, Jr, Baker LH. JAMA. 2011;306:1549–56. doi: 10.1001/jama.2011.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krehl S, Loewinger M, Florian S, Kipp AP, Banning A, Wessjohann LA, Brauer MN, Iori R, Esworthy RS, Chu FF, Brigelius-Flohe R. Carcinogenesis. 2012;33:620–8. doi: 10.1093/carcin/bgr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Science. 2003;300:1439–43. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- Kumar S, Bjornstedt M, Holmgren A. Eur J Biochem. 1992;207:435–39. doi: 10.1111/j.1432-1033.1992.tb17068.x. [DOI] [PubMed] [Google Scholar]
- Kumaraswamy E, Carlson BA, Morgan F, Miyoshi K, Robinson GW, Su D, Wang S, Southon E, Tessarollo L, Lee BJ, Gladyshev VN, Hennighausen L, Hatfield DL. Mol Cell Biol. 2003;23:1477–88. doi: 10.1128/MCB.23.5.1477-1488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaraswamy E, Malykh A, Korotkov KV, Kozyavkin S, Hu Y, Kwon SY, Moustafa ME, Carlson BA, Berry MJ, Lee BJ, Hatfield DL, Diamond AM, Gladyshev VN. J Biol Chem. 2000;275:35540–7. doi: 10.1074/jbc.M004014200. [DOI] [PubMed] [Google Scholar]
- Kurokawa S, Bellinger FP, Hill KE, Burk RF, Berry MJ. J Biol Chem. 2014a;289:9195–207. doi: 10.1074/jbc.M114.549014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa S, Eriksson S, Rose KL, Wu S, Motley AK, Hill S, Winfrey VP, McDonald WH, Capecchi MR, Atkins JF, Arner ES, Hill KE, Burk RF. Free Radic Biol Med. 2014b;69:67–76. doi: 10.1016/j.freeradbiomed.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa S, Hill KE, McDonald WH, Burk RF. J Biol Chem. 2012;287:28717–26. doi: 10.1074/jbc.M112.383521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X, Xing J, Gao H, Li S, Quan L, Jiang Y, Ding S, Xue Y. Biol Trace Elem Res. 2016 doi: 10.1007/s12011-016-0908-8. [DOI] [PubMed] [Google Scholar]
- Lance P, Alberts DS, Thompson PA, Fales L, Wang F, San Jose J, Jacobs ET, Goodman PJ, Darke AK, Yee M, Minasian L, Thompson IM, Roe DJ. Cancer Prev Res (Phila) 2017;10:45–54. doi: 10.1158/1940-6207.CAPR-16-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BJ, Rajagopalan M, Kim YS, You KH, Jacobson KB, Hatfield D. Mol Cell Biol. 1990;10:1940–9. doi: 10.1128/mcb.10.5.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Bar-Noy S, Kwon J, Levine RL, Stadtman TC, Rhee SG. Proc Natl Acad Sci U S A. 2000;97:2521–6. doi: 10.1073/pnas.050579797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Yang KS, Kwon J, Lee C, Jeong W, Rhee SG. J Biol Chem. 2002;277:20336–42. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- Leemhuis J, Bock HH. Commun Integr Biol. 2011;4:254–7. doi: 10.4161/cib.4.3.14890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z, Tian D, Zhang C, Zhao S, Su M. BMC Cancer. 2016;16:410. doi: 10.1186/s12885-016-2462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln DT, Ali Emadi EM, Tonissen KF, Clarke FM. Anticancer Res. 2003;23:2425–33. [PubMed] [Google Scholar]
- Lippman SM, Klein EA, Goodman PJ, Lucia MS, Thompson IM, Ford LG, Parnes HL, Minasian LM, Gaziano JM, Hartline JA, Parsons JK, Bearden JD, 3rd, Crawford ED, Goodman GE, Claudio J, Winquist E, Cook ED, Karp DD, Walther P, Lieber MM, Kristal AR, Darke AK, Arnold KB, Ganz PA, Santella RM, Albanes D, Taylor PR, Probstfield JL, Jagpal TJ, Crowley JJ, Meyskens FL, Jr, Baker LH, Coltman CA., Jr JAMA. 2009;301:39–51. doi: 10.1001/jama.2008.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Du J, Zhang Y, Sun W, Smith BJ, Oberley LW, Cullen JJ. Hum Gene Ther. 2006;17:105–16. doi: 10.1089/hum.2006.17.105. [DOI] [PubMed] [Google Scholar]
- Liu J, Hinkhouse MM, Sun W, Weydert CJ, Ritchie JM, Oberley LW, Cullen JJ. Hum Gene Ther. 2004;15:239–50. doi: 10.1089/104303404322886093. [DOI] [PubMed] [Google Scholar]
- Low SC, Grundner-Culemann E, Harney JW, Berry MJ. EMBO J. 2000;19:6882–90. doi: 10.1093/emboj/19.24.6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YP, Lou YR, Yen P, Newmark HL, Mirochnitchenko OI, Inouye M, Huang MT. Cancer Res. 1997;57:1468–74. [PubMed] [Google Scholar]
- Lubos E, Loscalzo J, Handy DE. Antioxid Redox Signal. 2011;15:1957–97. doi: 10.1089/ars.2010.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchman HA, Villemaire ML, Bismar TA, Carlson BA, Jirik FR. Am J Pathol. 2014;184:871–7. doi: 10.1016/j.ajpath.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorino M, Thomas JP, Girotti AW, Ursini F. Free Radic Res Commun. 1991;12–13(Pt 1):131–5. doi: 10.3109/10715769109145777. [DOI] [PubMed] [Google Scholar]
- Matsuzawa A, Ichijo H. Biochim Biophys Acta. 2008;1780:1325–36. doi: 10.1016/j.bbagen.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Meplan C, Dragsted LO, Ravn-Haren G, Tjonneland A, Vogel U, Hesketh J. PLoS One. 2013;8:e73316. doi: 10.1371/journal.pone.0073316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meplan C, Hughes DJ, Pardini B, Naccarati A, Soucek P, Vodickova L, Hlavata I, Vrana D, Vodicka P, Hesketh JE. Carcinogenesis. 2010;31:1074–9. doi: 10.1093/carcin/bgq076. [DOI] [PubMed] [Google Scholar]
- Meyer HA, Endermann T, Stephan C, Stoedter M, Behrends T, Wolff I, Jung K, Schomburg L. PLoS One. 2012;7:e46644. doi: 10.1371/journal.pone.0046644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Walker SW, Arthur JR, Nicol F, Pickard K, Lewin MH, Howie AF, Beckett GJ. Clin Sci (Lond) 2001;100:543–50. [PubMed] [Google Scholar]
- Moos PJ, Edes K, Cassidy P, Massuda E, Fitzpatrick FA. J Biol Chem. 2003;278:745–50. doi: 10.1074/jbc.M211134200. [DOI] [PubMed] [Google Scholar]
- Mostert V, Dreher I, Kohrle J, Abel J. FEBS Lett. 1999;460:23–6. doi: 10.1016/s0014-5793(99)01298-3. [DOI] [PubMed] [Google Scholar]
- Moustafa ME, Carlson BA, Anver MR, Bobe G, Zhong N, Ward JM, Perella CM, Hoffmann VJ, Rogers K, Combs GF, Jr, Schweizer U, Merlino G, Gladyshev VN, Hatfield DL. PLoS One. 2013;8:e57389. doi: 10.1371/journal.pone.0057389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa ME, Carlson BA, El-Saadani MA, Kryukov GV, Sun QA, Harney JW, Hill KE, Combs GF, Feigenbaum L, Mansur DB, Burk RF, Berry MJ, Diamond AM, Lee BJ, Gladyshev VN, Hatfield DL. Mol Cell Biol. 2001;21:3840–52. doi: 10.1128/MCB.21.11.3840-3852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MF, Florian S, Pommer S, Osterhoff M, Esworthy RS, Chu FF, Brigelius-Flohe R, Kipp AP. PLoS One. 2013;8:e72055. doi: 10.1371/journal.pone.0072055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawaki Y, Tsuchiya H, Kanbe T, Harada K, Yashima K, Nozaka K, Tanida O, Kohno M, Mukoyama T, Nishimuki E, Kojo H, Matsura T, Takahashi K, Osaki M, Ito H, Yodoi J, Murawaki Y, Shiota G. Cancer Lett. 2008;259:218–30. doi: 10.1016/j.canlet.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Naiki T, Naiki-Ito A, Asamoto M, Kawai N, Tozawa K, Etani T, Sato S, Suzuki S, Shirai T, Kohri K, Takahashi S. Carcinogenesis. 2014;35:1962–7. doi: 10.1093/carcin/bgu048. [DOI] [PubMed] [Google Scholar]
- Nasr MA, Fedele MJ, Esser K, Diamond AM. Free Radic Biol Med. 2004;37:187–95. doi: 10.1016/j.freeradbiomed.2004.04.038. [DOI] [PubMed] [Google Scholar]
- Olson GE, Winfrey VP, Nagdas SK, Hill KE, Burk RF. J Biol Chem. 2007;282:12290–7. doi: 10.1074/jbc.M611403200. [DOI] [PubMed] [Google Scholar]
- Outzen M, Tjonneland A, Larsen EH, Friis S, Larsen SB, Christensen J, Overvad K, Olsen A. Br J Nutr. 2016;115:1669–77. doi: 10.1017/S0007114516000726. [DOI] [PubMed] [Google Scholar]
- Papaioannou D, Cooper KL, Carroll C, Hind D, Squires H, Tappenden P, Logan RF. Colorectal Dis. 2011;13:1085–99. doi: 10.1111/j.1463-1318.2010.02289.x. [DOI] [PubMed] [Google Scholar]
- Pedersen MO, Hansen PB, Nielsen SL, Penkowa M. Leuk Lymphoma. 2010;51:314–28. doi: 10.3109/10428190903518329. [DOI] [PubMed] [Google Scholar]
- Pellatt AJ, Wolff RK, John EM, Torres-Mejia G, Hines LM, Baumgartner KB, Giuliano AR, Lundgreen A, Slattery ML. PLoS One. 2013;8:e80554. doi: 10.1371/journal.pone.0080554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosof L, Yerram S, Armstrong T, Chu N, Danilova L, Yanagisawa B, Hidalgo M, Azad N, Herman JG. Epigenetics. 2016 doi: 10.1080/15592294.2016.1265711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez S, Talens-Visconti R, Rius-Perez S, Finamor I, Sastre J. Free Radic Biol Med. 2017;104:75–103. doi: 10.1016/j.freeradbiomed.2016.12.048. [DOI] [PubMed] [Google Scholar]
- Peters U, Chatterjee N, Hayes RB, Schoen RE, Wang Y, Chanock SJ, Foster CB. Cancer Epidemiol Biomarkers Prev. 2008;17:1144–54. doi: 10.1158/1055-9965.EPI-07-2947. [DOI] [PubMed] [Google Scholar]
- Powis G, Mustacich D, Coon A. Free Radic Biol Med. 2000;29:312–22. doi: 10.1016/s0891-5849(00)00313-0. [DOI] [PubMed] [Google Scholar]
- Raninga PV, Di Trapani G, Vuckovic S, Bhatia M, Tonissen KF. Oncotarget. 2015;6:15410–24. doi: 10.18632/oncotarget.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raninga PV, Di Trapani G, Vuckovic S, Tonissen KF. Cell Cycle. 2016;15:559–72. doi: 10.1080/15384101.2015.1136038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves MA, Hoffmann PR. Cell Mol Life Sci. 2009;66:2457–78. doi: 10.1007/s00018-009-0032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards GM, Mehta MP. Expert Opin Pharmacother. 2007;8:351–9. doi: 10.1517/14656566.8.3.351. [DOI] [PubMed] [Google Scholar]
- Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, Shevde LA, Li W, Eschrich S, Daud A, Ju J, Matta J. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlof AK, Arner ES. Antioxid Redox Signal. 2004;6:41–52. doi: 10.1089/152308604771978336. [DOI] [PubMed] [Google Scholar]
- Saito Y, Hayashi T, Tanaka A, Watanabe Y, Suzuki M, Saito E, Takahashi K. J Biol Chem. 1999;274:2866–71. doi: 10.1074/jbc.274.5.2866. [DOI] [PubMed] [Google Scholar]
- Saito Y, Sato N, Hirashima M, Takebe G, Nagasawa S, Takahashi K. Biochem J. 2004;381:841–6. doi: 10.1042/BJ20040328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Fukushima N, Chang R, Matsubayashi H, Goggins M. Gastroenterology. 2006;130:548–65. doi: 10.1053/j.gastro.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Schoenmakers E, Carlson B, Agostini M, Moran C, Rajanayagam O, Bochukova E, Tobe R, Peat R, Gevers E, Muntoni F, Guicheney P, Schoenmakers N, Farooqi S, Lyons G, Hatfield D, Chatterjee K. J Clin Invest. 2016;126:992–6. doi: 10.1172/JCI84747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriever SC, Barnes KM, Evenson JK, Raines AM, Sunde RA. Exp Biol Med (Maywood) 2009;234:513–21. doi: 10.3181/0812-RM-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A, Lichti UF, Carlson BA, Cataisson C, Ryscavage AO, Mikulec C, Conrad M, Fischer SM, Hatfield DL, Yuspa SH. J Invest Dermatol. 2013;133:1731–41. doi: 10.1038/jid.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A, Lichti UF, Carlson BA, Ryscavage AO, Gladyshev VN, Yuspa SH, Hatfield DL. PLoS One. 2010;5:e12249. doi: 10.1371/journal.pone.0012249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedali A, Berry MJ. RNA. 2014;20:1248–56. doi: 10.1261/rna.043463.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki Y, Koizumi S, Ohno Y, Nagao T, Inoue K. Glia. 2006;54:606–18. doi: 10.1002/glia.20408. [DOI] [PubMed] [Google Scholar]
- Shrimali RK, Weaver JA, Miller GF, Starost MF, Carlson BA, Novoselov SV, Kumaraswamy E, Gladyshev VN, Hatfield DL. Neuromuscul Disord. 2007;17:135–42. doi: 10.1016/j.nmd.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, Eschrich S, Kis C, Levy S, Washington MK, Heslin MJ, Coffey RJ, Yeatman TJ, Shyr Y, Beauchamp RD. Gastroenterology. 2010;138:958–68. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg A, Sahaf B, Rosen A. Cancer Res. 2000;60:2281–9. [PubMed] [Google Scholar]
- Solinas G, Schiarea S, Liguori M, Fabbri M, Pesce S, Zammataro L, Pasqualini F, Nebuloni M, Chiabrando C, Mantovani A, Allavena P. J Immunol. 2010;185:642–52. doi: 10.4049/jimmunol.1000413. [DOI] [PubMed] [Google Scholar]
- Speckmann B, Pinto A, Winter M, Forster I, Sies H, Steinbrenner H. Free Radic Biol Med. 2010;49:777–85. doi: 10.1016/j.freeradbiomed.2010.05.035. [DOI] [PubMed] [Google Scholar]
- Stadtman TC. Annu Rev Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- Stadtman TC. Ann N Y Acad Sci. 2000;899:399–402. doi: 10.1111/j.1749-6632.2000.tb06203.x. [DOI] [PubMed] [Google Scholar]
- Steinbrecher A, Meplan C, Hesketh J, Schomburg L, Endermann T, Jansen E, Akesson B, Rohrmann S, Linseisen J. Cancer Epidemiol Biomarkers Prev. 2010;19:2958–68. doi: 10.1158/1055-9965.EPI-10-0364. [DOI] [PubMed] [Google Scholar]
- Streicher KL, Sylte MJ, Johnson SE, Sordillo LM. Nutr Cancer. 2004;50:221–31. doi: 10.1207/s15327914nc5002_13. [DOI] [PubMed] [Google Scholar]
- Su D, Novoselov SV, Sun QA, Moustafa ME, Zhou Y, Oko R, Hatfield DL, Gladyshev VN. J Biol Chem. 2005;280:26491–8. doi: 10.1074/jbc.M503638200. [DOI] [PubMed] [Google Scholar]
- Sun QA, Su D, Novoselov SV, Carlson BA, Hatfield DL, Gladyshev VN. Biochemistry. 2005;44:14528–37. doi: 10.1021/bi051321w. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Pitchakarn P, Ogawa K, Naiki-Ito A, Chewonarin T, Punfa W, Asamoto M, Shirai T, Takahashi S. Toxicology. 2013;311:115–23. doi: 10.1016/j.tox.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Kelly VP, Motohashi H, Nakajima O, Takahashi S, Nishimura S, Yamamoto M. J Biol Chem. 2008;283:2021–30. doi: 10.1074/jbc.M708352200. [DOI] [PubMed] [Google Scholar]
- Tsuji PA, Carlson BA, Naranjo-Suarez S, Yoo MH, Xu XM, Fomenko DE, Gladyshev VN, Hatfield DL, Davis CD. PLoS One. 2012;7:e50574. doi: 10.1371/journal.pone.0050574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji PA, Naranjo-Suarez S, Carlson BA, Tobe R, Yoo MH, Davis CD. Nutrients. 2011;3:805–17. doi: 10.3390/nu3090805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursini F, Maiorino M, Valente M, Ferri L, Gregolin C. Biochim Biophys Acta. 1982;710:197–211. doi: 10.1016/0005-2760(82)90150-3. [DOI] [PubMed] [Google Scholar]
- van Rensburg SJ, Hall JM, Gathercole PS. Nutr Cancer. 1986;8:163–70. doi: 10.1080/01635588609513890. [DOI] [PubMed] [Google Scholar]
- Walczak R, Westhof E, Carbon P, Krol A. RNA. 1996;2:367–79. [PMC free article] [PubMed] [Google Scholar]
- Wallace K, Byers T, Morris JS, Cole BF, Greenberg ER, Baron JA, Gudino A, Spate V, Karagas MR. Cancer Epidemiol Biomarkers Prev. 2003;12:464–7. [PubMed] [Google Scholar]
- Wang H, Luo K, Tan LZ, Ren BG, Gu LQ, Michalopoulos G, Luo JH, Yu YP. J Biol Chem. 2012;287:16890–902. doi: 10.1074/jbc.M111.322636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Bonorden MJ, Li GX, Lee HJ, Hu H, Zhang Y, Liao JD, Cleary MP, Lu J. Cancer Prev Res (Phila) 2009a;2:484–95. doi: 10.1158/1940-6207.CAPR-08-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Gong L, Dong R, Qiao Q, He XL, Chu YK, Du XL, Yang Y, Zang L, Nan J, Lin C, Lu JG. J Int Med Res. 2009b;37:169–74. doi: 10.1177/147323000903700120. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang J, Xu T. Eur J Pharmacol. 2008;579:66–73. doi: 10.1016/j.ejphar.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Whitin JC, Bhamre S, Tham DM, Cohen HJ. Am J Physiol Renal Physiol. 2002;283:F20–8. doi: 10.1152/ajprenal.00014.2001. [DOI] [PubMed] [Google Scholar]
- Woutersen RA, Appel MJ, Van Garderen-Hoetmer A. Food Chem Toxicol. 1999;37:981–4. doi: 10.1016/s0278-6915(99)00093-9. [DOI] [PubMed] [Google Scholar]
- Xu XM, Turanov AA, Carlson BA, Yoo MH, Everley RA, Nandakumar R, Sorokina I, Gygi SP, Gladyshev VN, Hatfield DL. Proc Natl Acad Sci U S A. 2010;107:21430–4. doi: 10.1073/pnas.1009947107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Diamond AM. Biomark Res. 2013;1:15. doi: 10.1186/2050-7771-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Shen Y, Wei J, Liu F. Oncotarget. 2015;6:22006–27. doi: 10.18632/oncotarget.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, Brown LM, Girotti AW, Cornish VW, Schreiber SL, Stockwell BR. Cell. 2014;156:317–31. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant LJ, Ran Q, Rao L, Van Remmen H, Shibatani T, Belter JG, Motta L, Richardson A, Prolla TA. Free Radic Biol Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- Yao Y, Pei F, Kang P. Nutrition. 2011;27:1095–100. doi: 10.1016/j.nut.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Yi YS, Park SG, Byeon SM, Kwon YG, Jung G. Biochim Biophys Acta. 2003;1638:249–56. doi: 10.1016/s0925-4439(03)00090-5. [DOI] [PubMed] [Google Scholar]
- Yoo MH, Xu XM, Carlson BA, Gladyshev VN, Hatfield DL. J Biol Chem. 2006;281:13005–8. doi: 10.1074/jbc.C600012200. [DOI] [PubMed] [Google Scholar]
- Yoo SE, Chen L, Na R, Liu Y, Rios C, Van Remmen H, Richardson A, Ran Q. Free Radic Biol Med. 2012;52:1820–7. doi: 10.1016/j.freeradbiomed.2012.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YP, Yu G, Tseng G, Cieply K, Nelson J, Defrances M, Zarnegar R, Michalopoulos G, Luo JH. Cancer Res. 2007;67:8043–50. doi: 10.1158/0008-5472.CAN-07-0648. [DOI] [PubMed] [Google Scholar]
- Zhang S, Rocourt C, Cheng WH. Mech Ageing Dev. 2010;131:253–60. doi: 10.1016/j.mad.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Zhu S, Zhang Q, Sun X, Zeh HJ, Lotze MT, Kang R, Tang D. Cancer Res. 2017 doi: 10.1158/0008-5472.CAN-16-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]