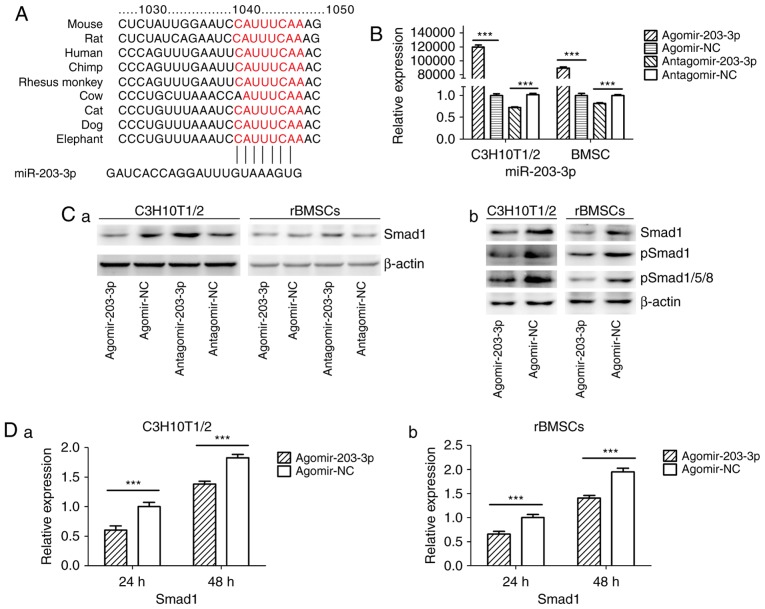
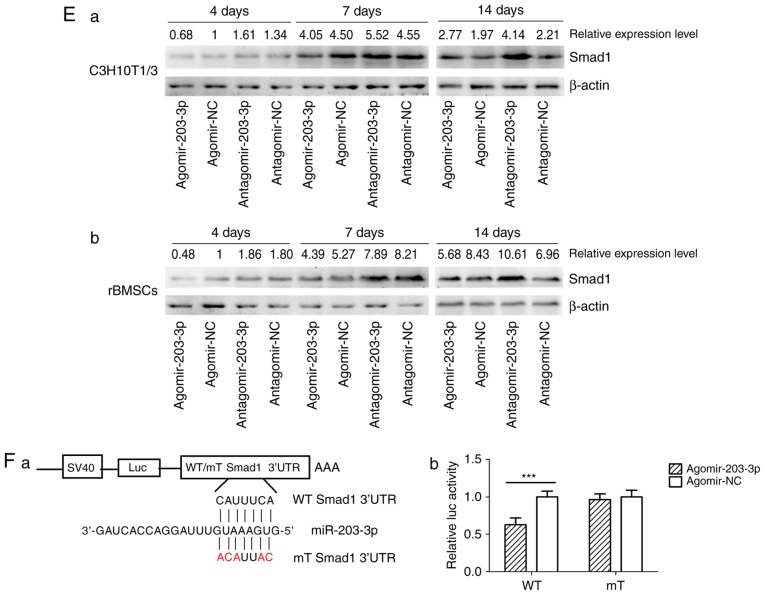
Figure 4.
miR-203-3p targeting the Smad1 gene. (A) A putative target site of miR-203-3p, which is highly conserved in mammals, was predicted to be located in the 3′-UTR of Smad1 mRNA, by using TargetScan software. The numbers represent the position of the 'seed region' matching miR-203-3p within the 3′-UTR sequences. (B) Reverse transcription-quantitative polymerase chain reaction analysis of levels of miR-203-3p expression in C3H10T1/2 cells and rBMSCs transfected with agomir-203-3p, agomir-NC, antagomir-203-3p and antagomir-NC. (C) Protein was extracted at 48 h after transfection. (a) Western blot analysis demonstrated that the protein levels of Smad1 were repressed by overexpression of miR-203-3p and promoted by inhibition of miR-203-3p. (b) Expression of bone morphogenetic protein/Smad pathway-associated proteins in C3H10T1/2 cells and rBMSCs. Representative western blot images demonstrate that Smad1, p-Smad1 and p-Smad1/5/8 levels are repressed by overexpression of miR-203-3p. (D) mRNA was extracted at 24 and 48 h after transfection. The mRNA expression of Smad1 was depressed by overexpression of miR-203-3p in (a) C3H10T1/2 cells and (b) rBMSCs. (E) Smad1 protein levels in (a) C3H10T1/2 cells and (b) rBMSCs transfected with agomir-203-3p, agomir-NC, antagomir-203-3p and antagomir-NC on the days 4, 7 and 14 of osteogenic induction was detected by western blot analysis, revealing that overexpression of miR-203-3p caused the expression of Smad1 to be (a) depressed and retarded in C3H10T1/2 cells and (b) to be depressed in rBMSCs. Overexpression of miR-203-3p suppressed the expression of Smad1 in (a) C3H10T1/2 cells and (b) rBMSCs, while inhibition of miR-203-3p resulted in the promotion of Smad1. Agomir delayed the expression of Smad1 so that, in the osteogenic process, the Agomir group demonstrated the highest expression levels of Smad1 later than the Agomir-NC group. At day 14, Smad1 appeared to be increased in the Agomir group compared with that observed in the Agomir-NC group. (F-a) A luciferase reporter system containing a binding site (Smad1-3′-UTR-WT) or a mutated site (Smad1-3′-UTR-mut) located in the downstream region of the pGL3 luciferase reporter gene was constructed. C3H10T1/2 cells were co-transfected with pGL3-Smad1 or pGL3-Smad1-mut, phRL-null and Agomir-203-3p or Agomir-NC. (b) Compared with the negative control, the luciferase activity in the group co-transfected with Agomir-203-3p and pGL3-Smad1-WT was decreased by 37.3%, while Agomir-203-3p did not affect the luciferase activity of the pGL3-Smad1-mut vector. Values are expressed as the mean ± standard deviation (n=3). ***P<0.001. miR, microRNA; NC, negative control; Agomir, miR agonist; Antagomir, miR antagonist; UTR, untranslated region; mut/Mt, mutant; WT, wild-type; rBMSCs, rat bone marrow mesenchymal stem cells; pSmad, phosphorylated Smad; Luc, luciferase.