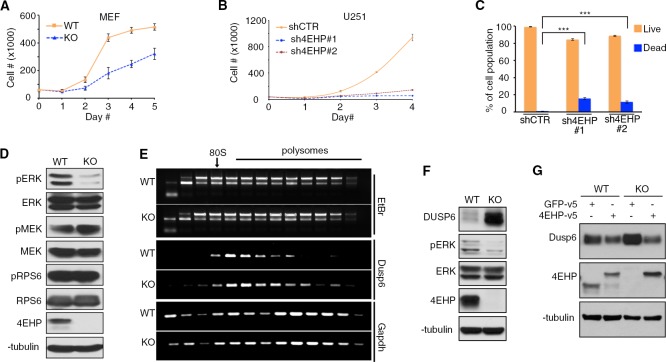
Figure 2. Depletion of 4EHP expression affects cell proliferation, survival, and ERK1/2 phosphorylation.
(A) Cell proliferation assay. WT and 4EHP-KO MEFs were seeded in 6-well plates and trypsinized after the indicated time points and cell numbers determined using a hematocytometer. Data are mean ± SD (n = 3). (B) Cell proliferation assay. U251 cells with stable expression of shCTR (control), sh4EHP#1, and sh4EHP#2 were seeded in 6-well plates. Cells were trypsinized after the indicated time points and cell numbers determined using a hematocytometer. Data are mean ± SD (n = 3). (C) Quantitation of cell death by FACS assay; Sub-G population was considered as ‘Dead’ and G0/1, S and G2/M population was combined as ‘Live’. Data are mean ± SD (n = 3). (D) WB for the indicated proteins in the WT and 4EHP-KO MEFs. (E) Polysome profiling/RT-PCR; RNA was extracted from each fraction (collected as described in Figure 2—figure supplement 1J), subjected to electrophoresis on agarose gel and visualized, using Ethidium Bromide (EtBr) staining. RT-PCR analyses of total RNA in each fraction was carried out with primers specific for Dusp6 and Gapdh mRNAs. (F) WB on the indicated proteins in WT and 4EHP-KO MEFs. (G) WB for the indicated proteins in the WT and 4EHP-KO MEFs, expressing a v5-tagged GFP (GFP-v5) or v5-tagged 4EHP (4EHP-v5).