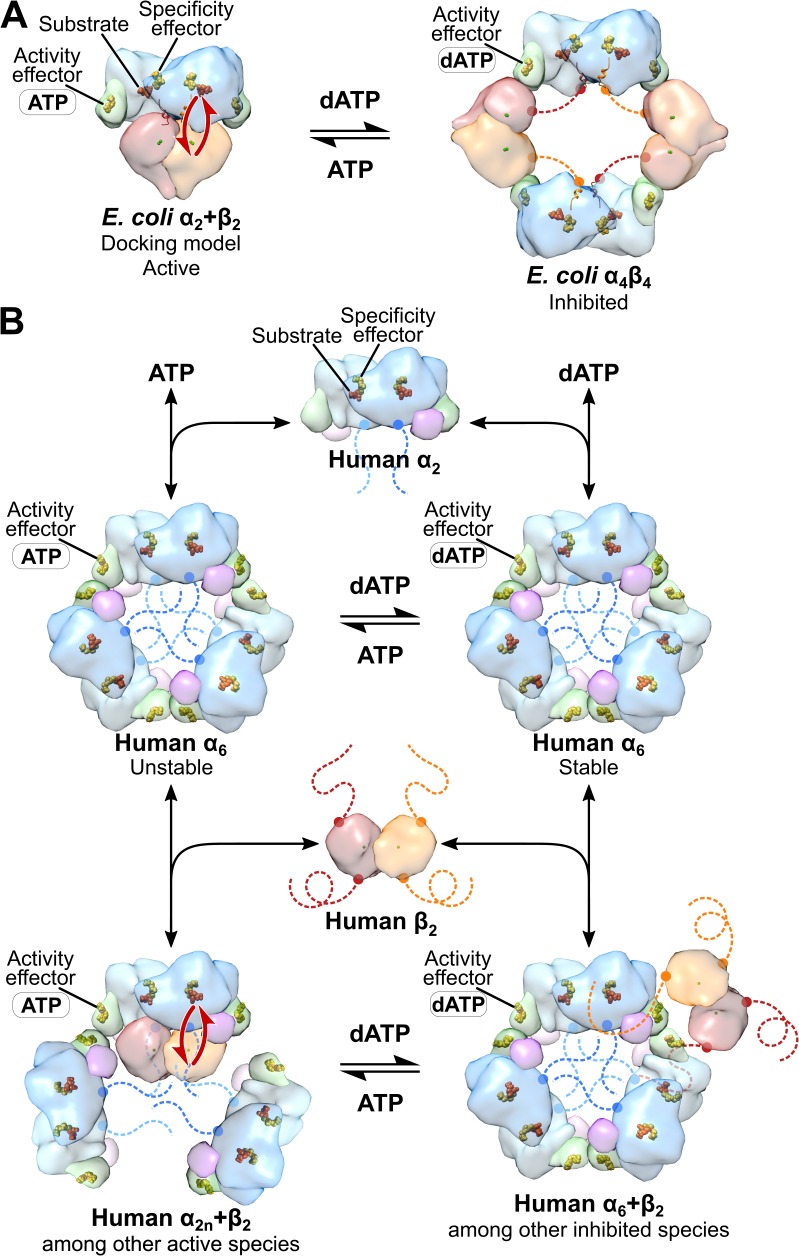
Figure 2. Comparison of mechanisms of allosteric regulation of activity for E. coli and human RNRs.
(A) The molecular basis of allosteric regulation of activity in class Ia RNR from E. coli. dATP inhibits activity by binding to the activity effector site in the cone domain (green) in α and promoting conversion of the active α2β2 complex (modeled by docking PDB: 5CNS chains A and B with PDB: 1RIB) to an inhibited α4β4 ring (PDB: 5CNS). In particular, with dATP bound to the activity effector site in α, the cone domain forms an interface with β, leading to α4β4 ring formation. ATP restores activity by displacing dATP in the activity effector site in the cone domain, which disrupts the α-β interface of the α4β4 ring, and further pushes the equilibrium toward the α2β2 complex. The molecular basis by which dATP promotes α-β interface formation and ATP disrupts it has not been established. Long-distance radical transfer between α and β is illustrated by red arrows. (B) The molecular basis of allosteric regulation of activity in human class Ia RNR (see text in Discussion). Briefly, α2 forms α6 in the presence of both ATP and dATP. The stability of the hexamer formed determines whether the enzyme is active when β2 is added. dATP-induced hexamers are stable and inactive whereas ATP-induced hexamers are unstable and activated by β2 addition. Schematic of human RNR was prepared using the same models and coloring as Figure 1.